Team:Heidelberg/Notebook/miRNA Kit/September
From 2010.igem.org
| Line 2: | Line 2: | ||
{{:Team:Heidelberg/Pagetop|note_mirna_kit}} | {{:Team:Heidelberg/Pagetop|note_mirna_kit}} | ||
| - | = 01/09/2010 = | + | ===01/09/2010=== |
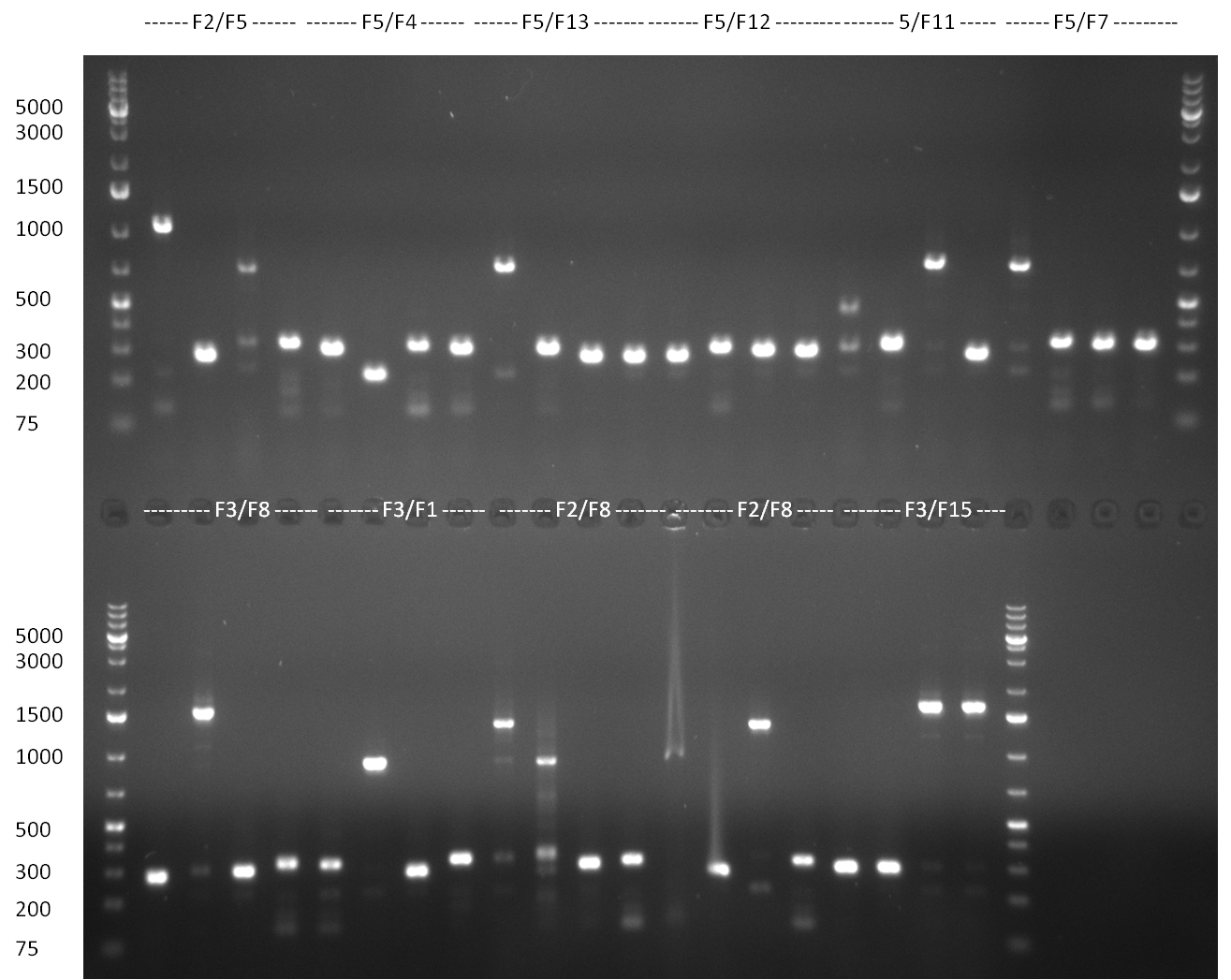
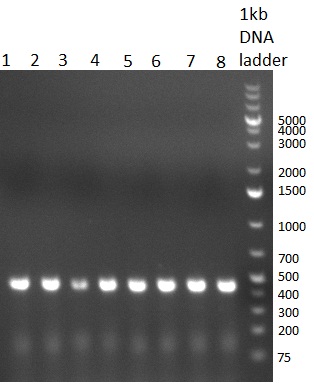
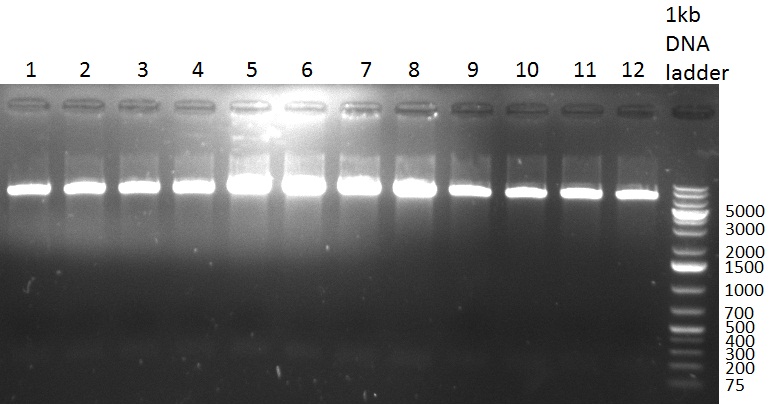
[[Image:20100901_gel1bb.png|thumb|350 px|right|Gel 100901-1]] | [[Image:20100901_gel1bb.png|thumb|350 px|right|Gel 100901-1]] | ||
[[Image:20100901_gel2bb.png|thumb|350 px|right|Gel 100901-2]] | [[Image:20100901_gel2bb.png|thumb|350 px|right|Gel 100901-2]] | ||
| Line 9: | Line 9: | ||
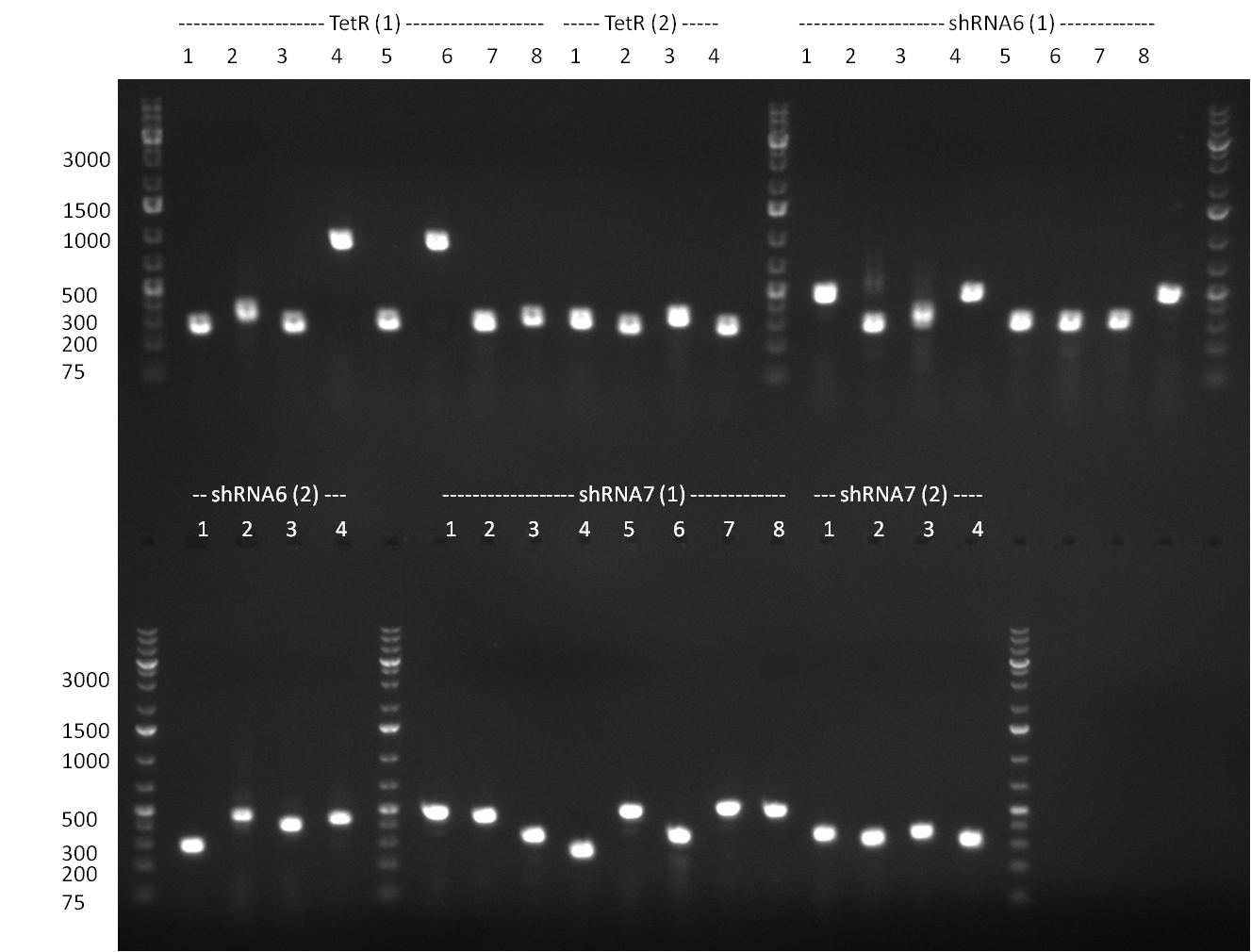
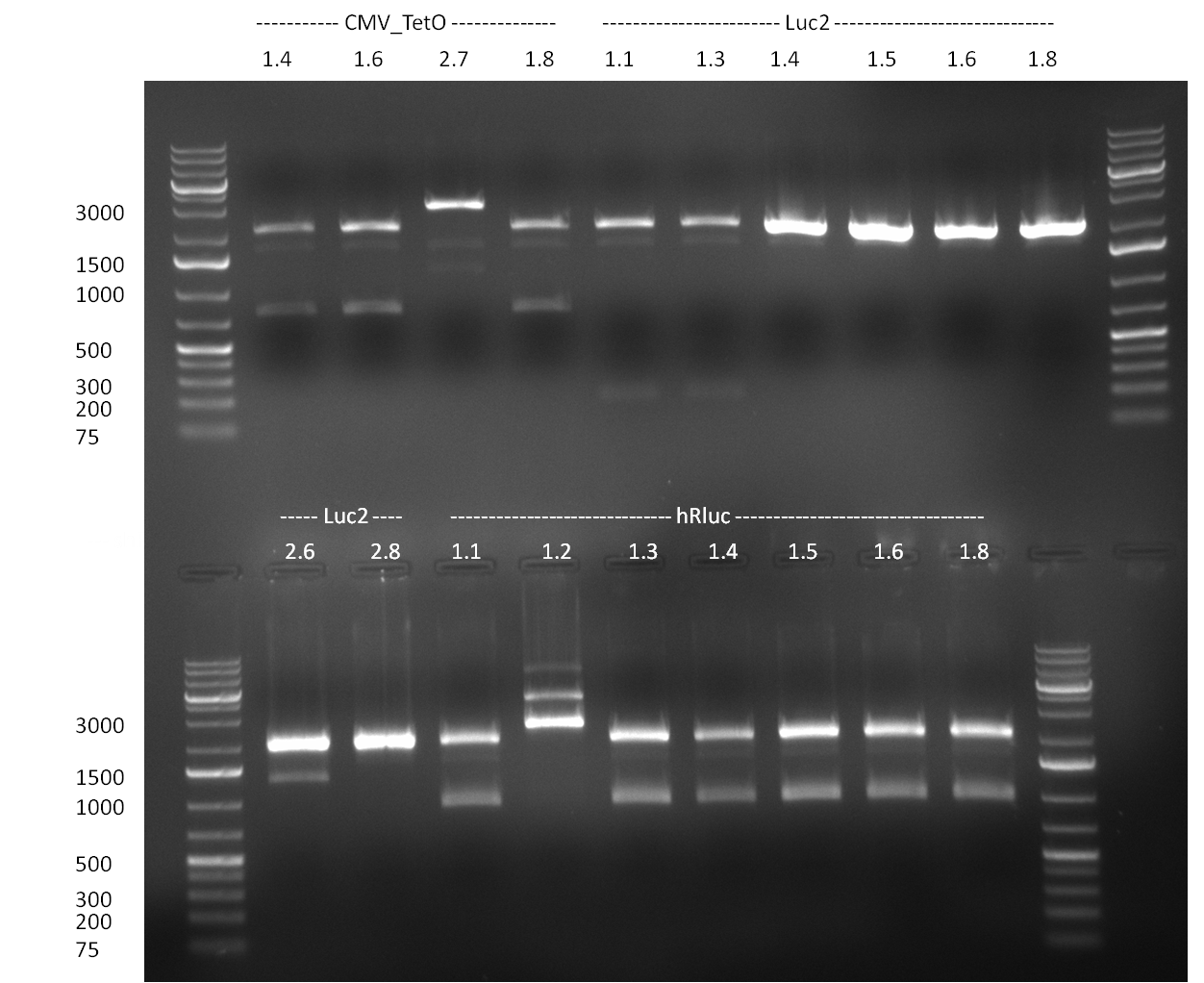
* PCR for the construction of the standardized Tet Repressor was performed. The PCR was set up using the Phusion HF MasterMix and standard PCR protocol was applied (annealing temperature 59 °C, 2 replicates). The result of the PCR was analyzed on a 1 % agarose gel (gel 100901-2, lower right side) and the right 700 bp band was clearly visible. | * PCR for the construction of the standardized Tet Repressor was performed. The PCR was set up using the Phusion HF MasterMix and standard PCR protocol was applied (annealing temperature 59 °C, 2 replicates). The result of the PCR was analyzed on a 1 % agarose gel (gel 100901-2, lower right side) and the right 700 bp band was clearly visible. | ||
* Digestion of the TetR fragment with EcoRI/PstI was performed according to the standard digestion protocol and the fragment was subsequently ligated into pSB1C3 (SAP treated or non-treated). The ligation mix was transformed into Top10 cells. | * Digestion of the TetR fragment with EcoRI/PstI was performed according to the standard digestion protocol and the fragment was subsequently ligated into pSB1C3 (SAP treated or non-treated). The ligation mix was transformed into Top10 cells. | ||
| - | + | <br /> | |
| - | = 02/09/2010 = | + | <br /> |
| + | ===02/09/2010=== | ||
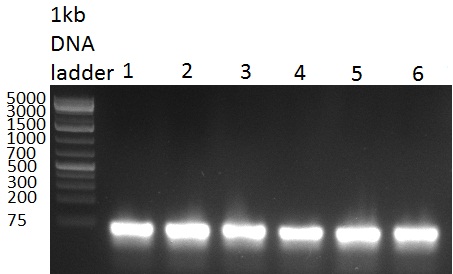
[[Image:20100802_gel1.png|thumb|350 px|right|Gel 100902-1]] | [[Image:20100802_gel1.png|thumb|350 px|right|Gel 100902-1]] | ||
<br /> | <br /> | ||
| Line 30: | Line 31: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
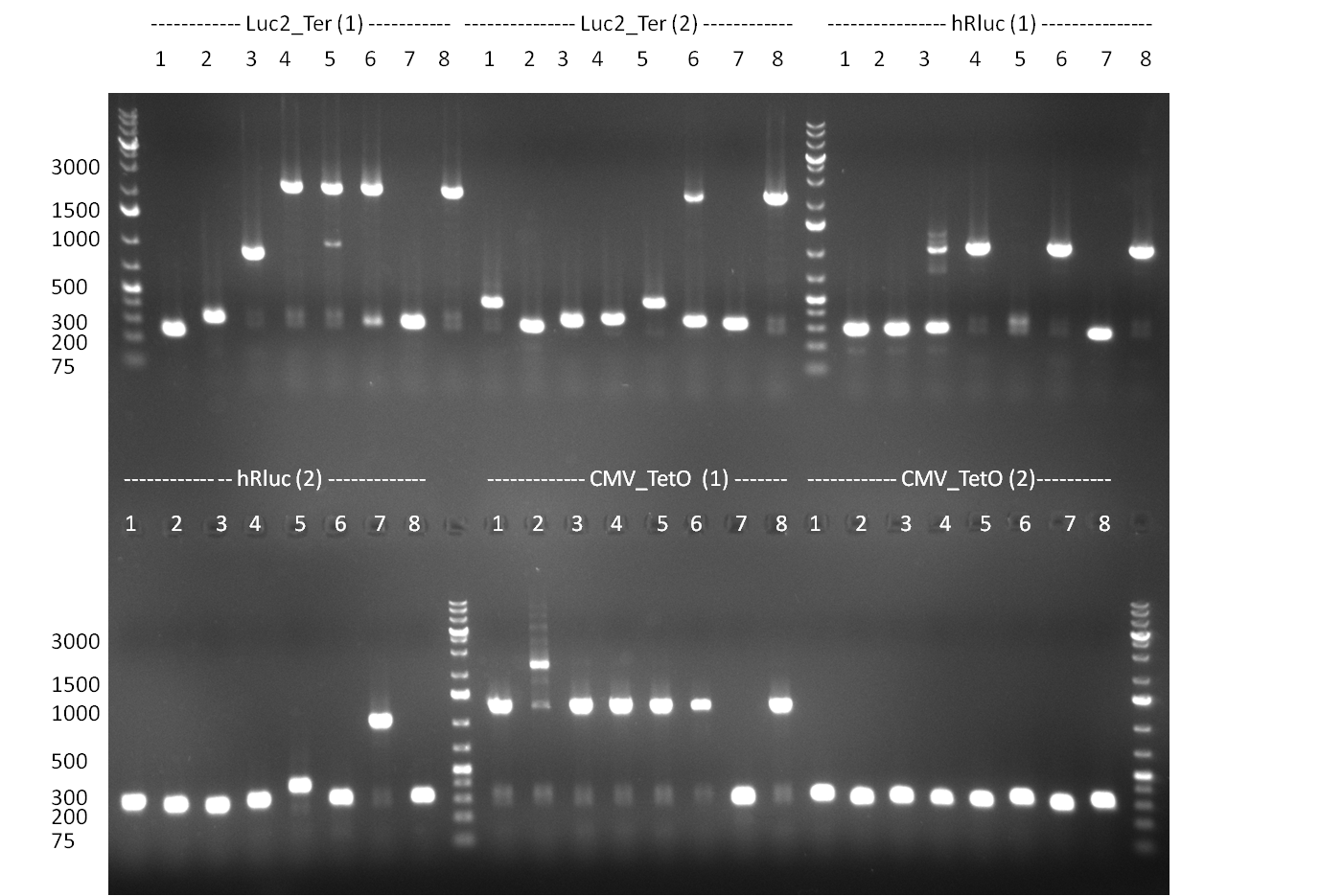
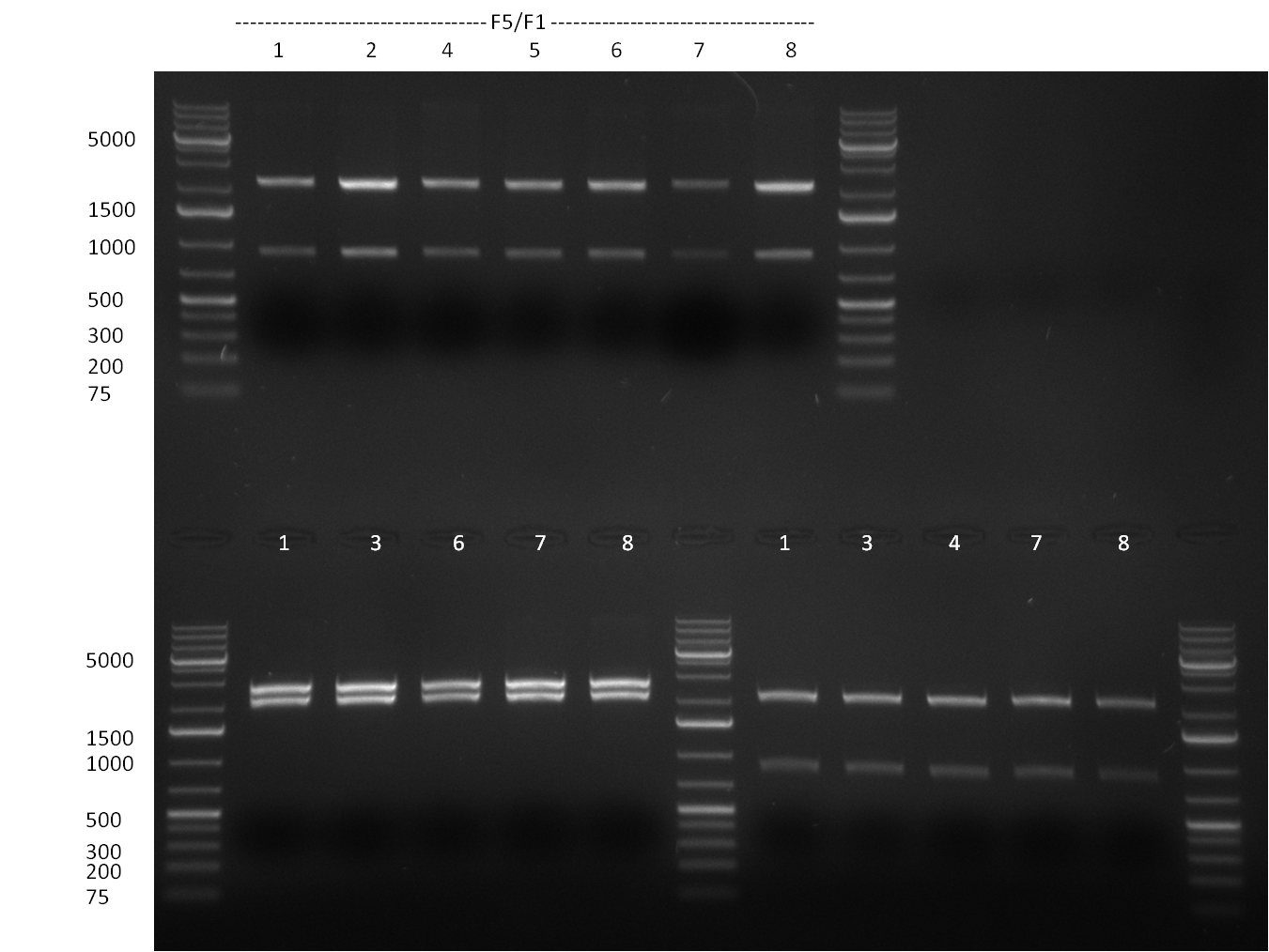
| - | = 03/09/2010 = | + | ===03/09/2010=== |
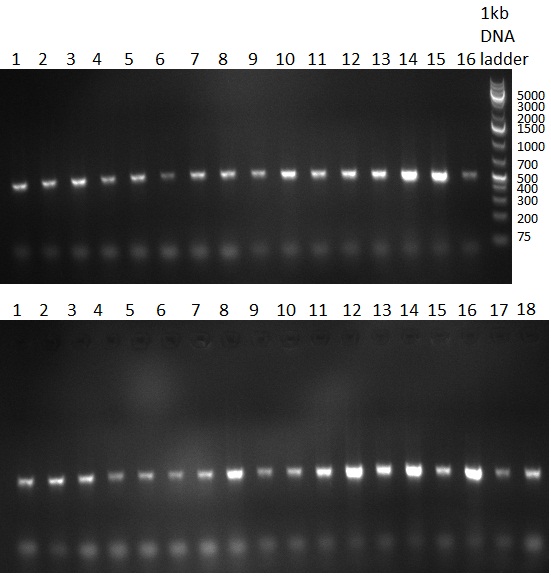
[[Image:20100903_gel1.png|thumb|350 px|right|Gel 100903-1]] | [[Image:20100903_gel1.png|thumb|350 px|right|Gel 100903-1]] | ||
[[Image:20100903_gel3.png|thumb|350 px|right|Gel 100903-2]] | [[Image:20100903_gel3.png|thumb|350 px|right|Gel 100903-2]] | ||
| Line 40: | Line 41: | ||
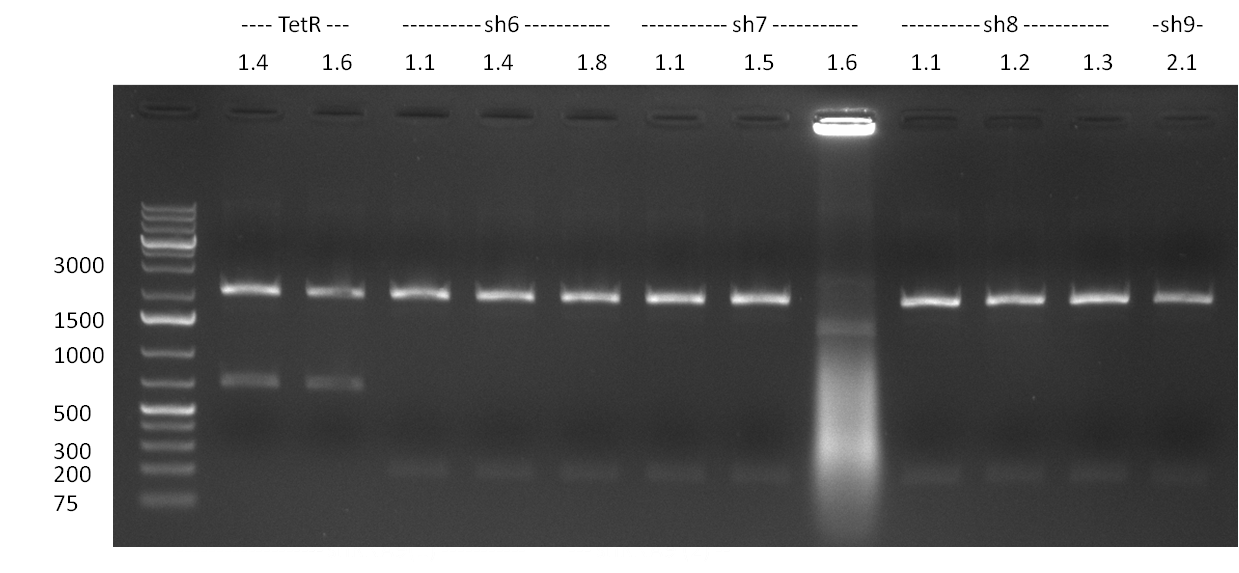
* cloning of shRNAs into pcDNA5/FRT/TO | * cloning of shRNAs into pcDNA5/FRT/TO | ||
| - | = 04/09/2010 = | + | ===04/09/2010=== |
* mini-prep of pcDNA5 with shRNAs 1, 2, 3, 5, 5<nowiki>*</nowiki> and 6 (each four colonies from two independent transformations) | * mini-prep of pcDNA5 with shRNAs 1, 2, 3, 5, 5<nowiki>*</nowiki> and 6 (each four colonies from two independent transformations) | ||
| Line 57: | Line 58: | ||
| - | = 05/09/2010 = | + | ===05/09/2010=== |
<br /> | <br /> | ||
* transformation of final Kit constructs F1 - F16 into E. coli Top10 cells | * transformation of final Kit constructs F1 - F16 into E. coli Top10 cells | ||
| Line 71: | Line 72: | ||
<br /> | <br /> | ||
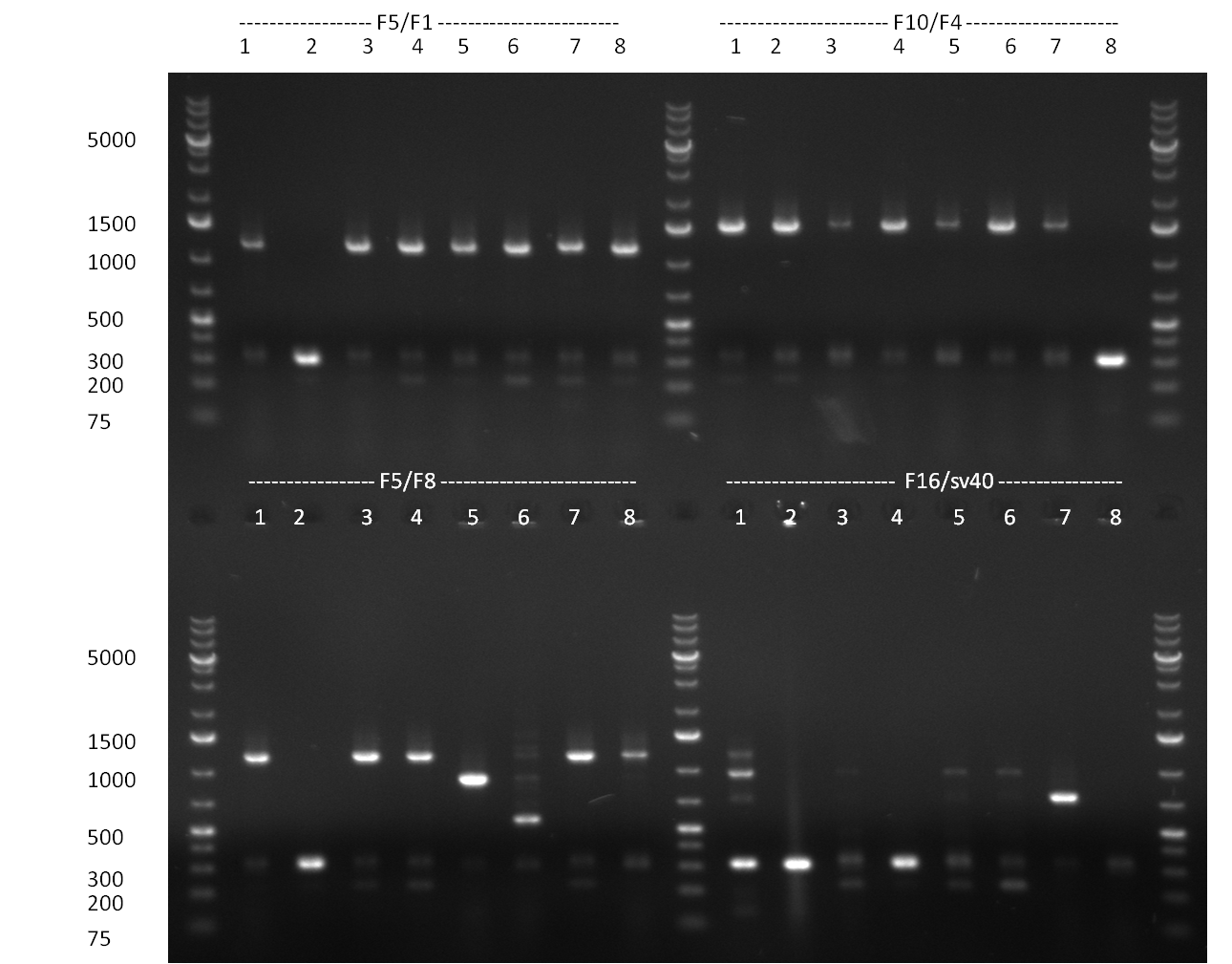
| - | = 07/09/2010 = | + | ===07/09/2010=== |
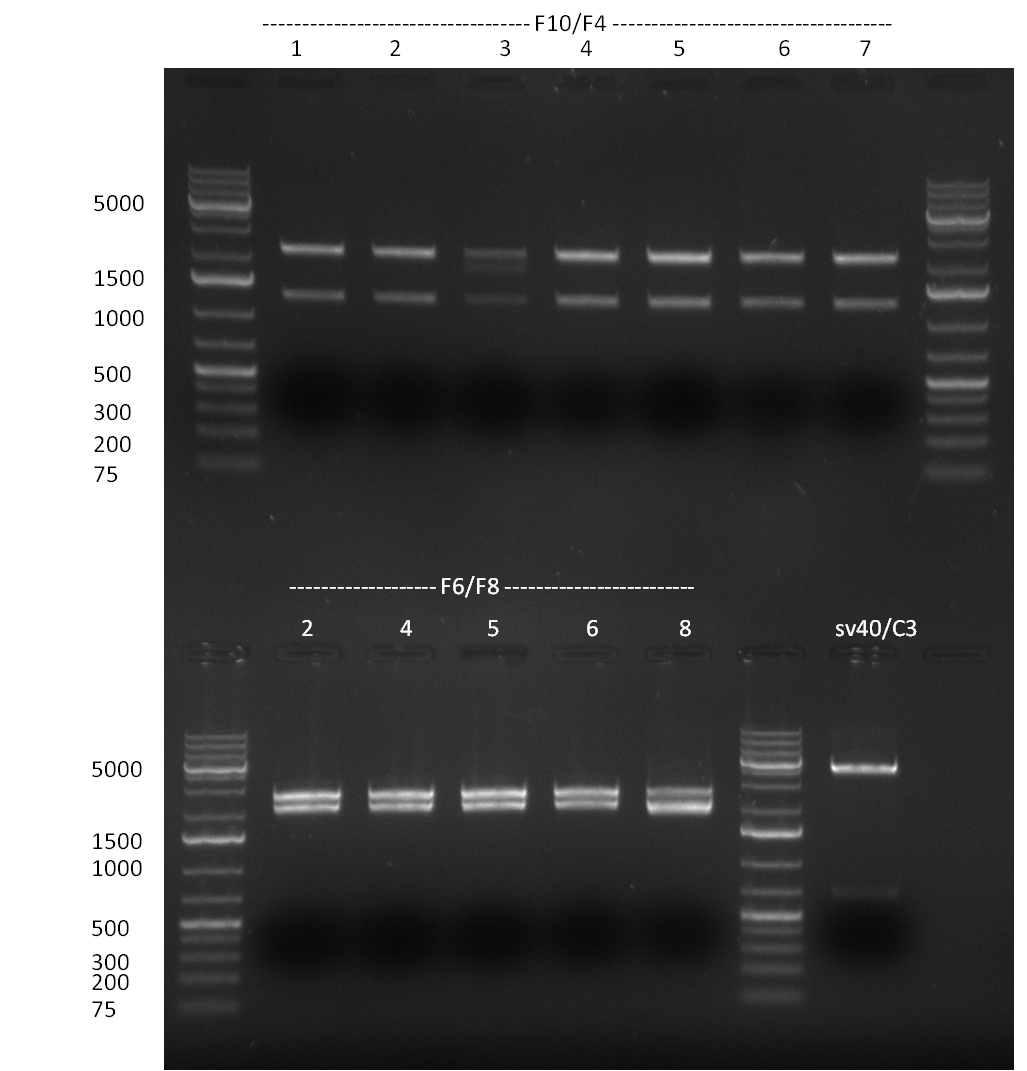
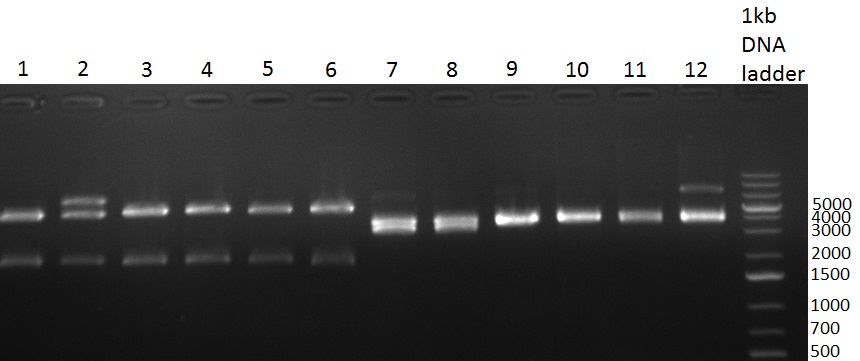
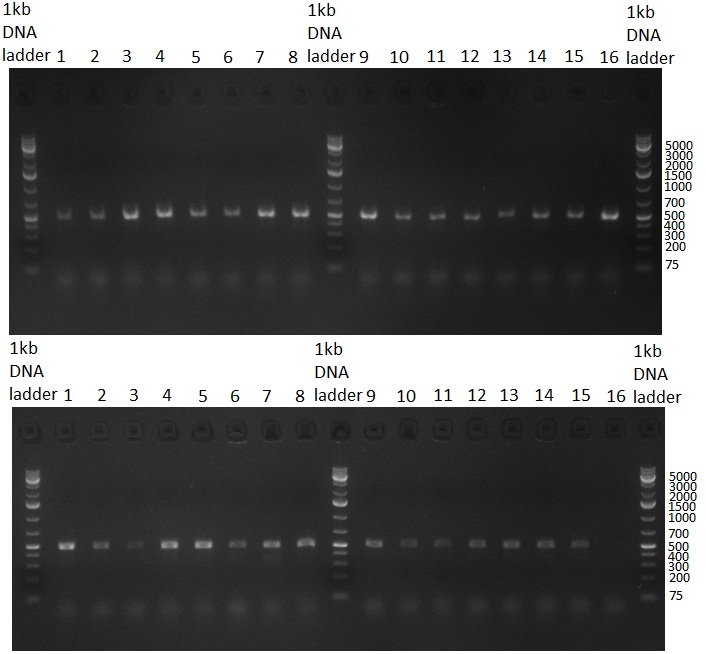
[[Image:20100908_gel1.png|thumb|350 px|right|Gel 100907-1: Ligation Control. Numbers indicate the F-Construct numbers of the parts assembled]] | [[Image:20100908_gel1.png|thumb|350 px|right|Gel 100907-1: Ligation Control. Numbers indicate the F-Construct numbers of the parts assembled]] | ||
<br /> | <br /> | ||
| Line 93: | Line 94: | ||
<br /> | <br /> | ||
| - | = 08/09/2010 = | + | ===08/09/2010=== |
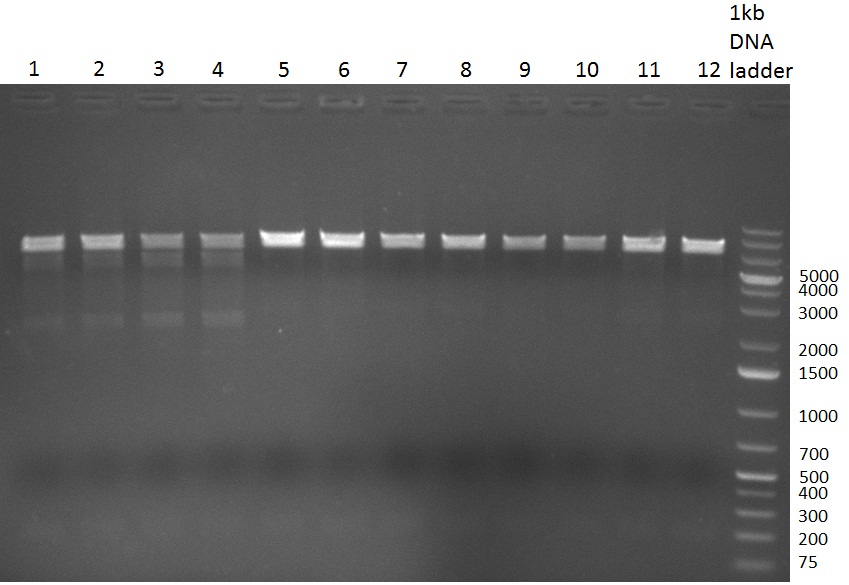
[[Image:20100908_gel2.png|thumb|350 px|right|Gel 100908-1: Colony-PCR.]] | [[Image:20100908_gel2.png|thumb|350 px|right|Gel 100908-1: Colony-PCR.]] | ||
<br /> | <br /> | ||
| Line 106: | Line 107: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
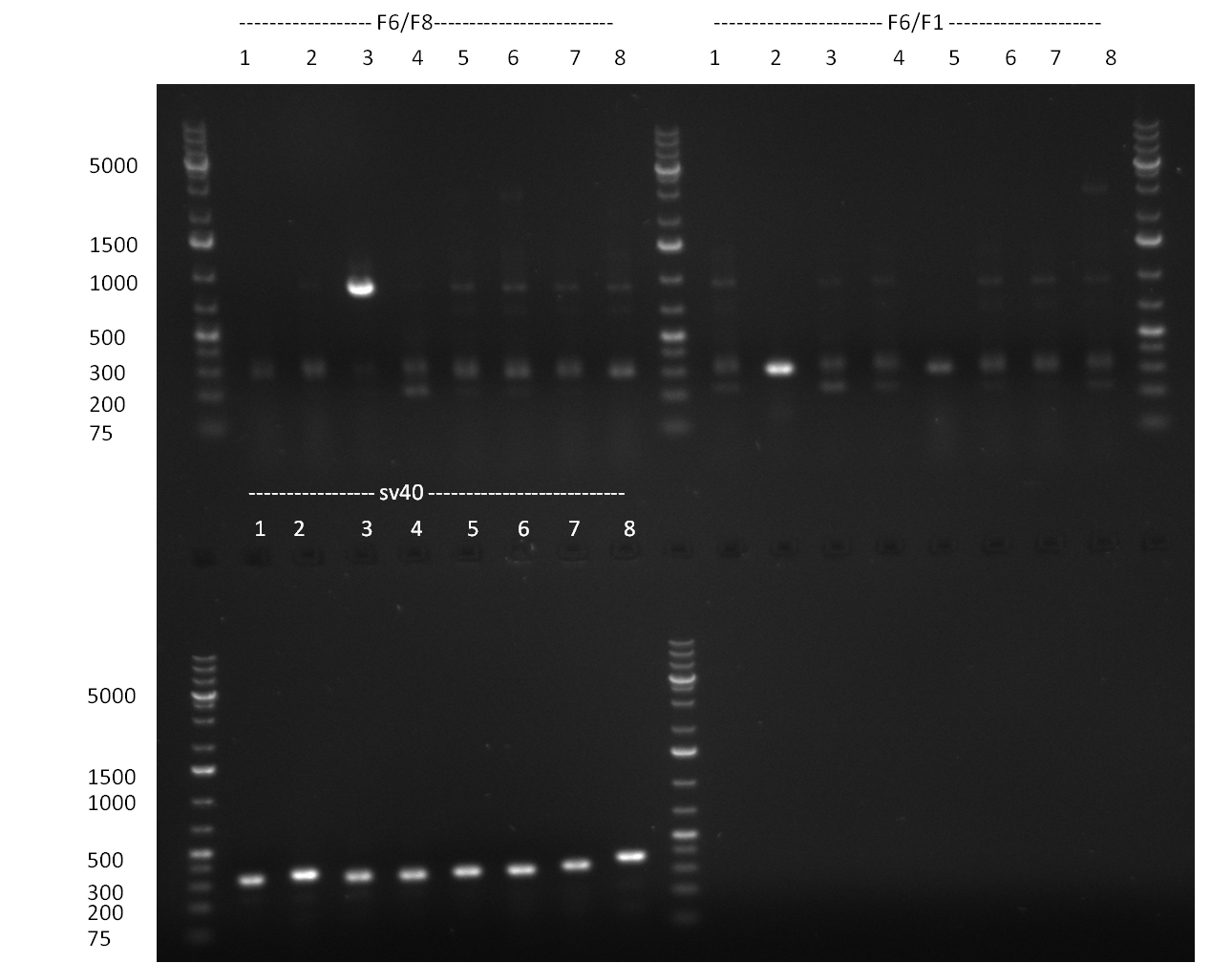
| - | = 09/09/2010 = | + | ===09/09/2010=== |
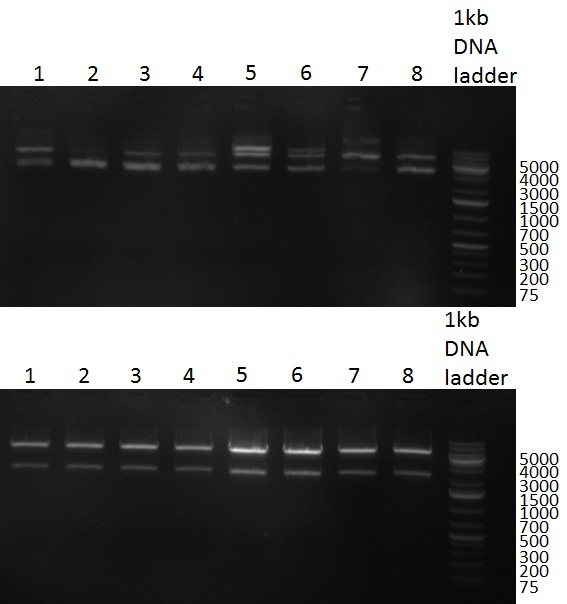
[[Image:20100909_gel1.png|thumb|350 px|right|Gel 100909-1]] | [[Image:20100909_gel1.png|thumb|350 px|right|Gel 100909-1]] | ||
[[Image:20100909_F6_negative_positive_sm.png |thumb|350 px|right|Gel 100909-2]] | [[Image:20100909_F6_negative_positive_sm.png |thumb|350 px|right|Gel 100909-2]] | ||
| Line 124: | Line 125: | ||
<br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /> | ||
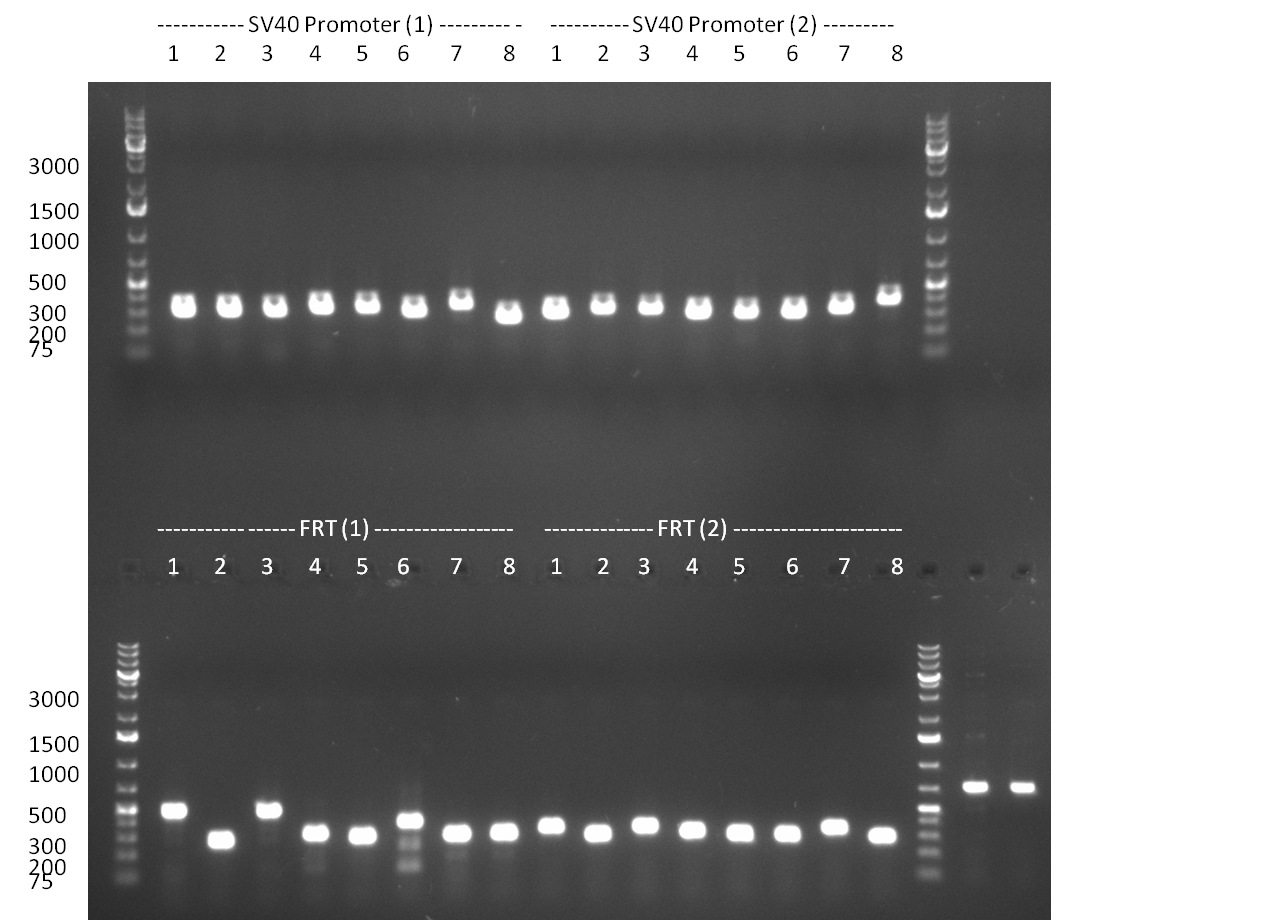
| - | = 10/09/2010 = | + | ===10/09/2010=== |
<br /> | <br /> | ||
* PCR of SV40 Terminator with BamHI site according to standard PCR protocol (annealing @ 58 °C) | * PCR of SV40 Terminator with BamHI site according to standard PCR protocol (annealing @ 58 °C) | ||
| Line 143: | Line 144: | ||
| - | = 11/09/2010 = | + | ===11/09/2010=== |
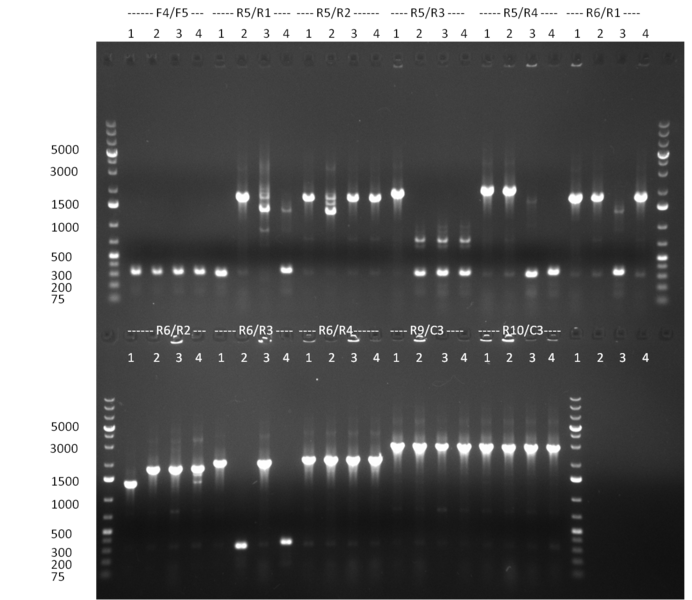
[[Image:20100911_gel1.png|thumb|350 px|right|Gel 100911-1]] | [[Image:20100911_gel1.png|thumb|350 px|right|Gel 100911-1]] | ||
[[Image:20100911_gel2.png|thumb|350 px|right|Gel 100911-2]] | [[Image:20100911_gel2.png|thumb|350 px|right|Gel 100911-2]] | ||
| Line 155: | Line 156: | ||
| - | = 12/09/2010 = | + | ===12/09/2010=== |
[[Image:20100912_gel1.png|thumb|350 px|right|Gel 100912-1]] | [[Image:20100912_gel1.png|thumb|350 px|right|Gel 100912-1]] | ||
[[Image:20100912_gel2.png|thumb|350 px|right|Gel 100912-2]] | [[Image:20100912_gel2.png|thumb|350 px|right|Gel 100912-2]] | ||
| Line 165: | Line 166: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| - | = 13/09/2010 = | + | ===13/09/2010=== |
<br /> | <br /> | ||
* digestion of the following parts: | * digestion of the following parts: | ||
| Line 182: | Line 183: | ||
| - | = 14/09/2010 = | + | ===14/09/2010=== |
[[Image:20100914_HD10.png|thumb|350 px|right|Gel 100914-1]] | [[Image:20100914_HD10.png|thumb|350 px|right|Gel 100914-1]] | ||
[[Image:20100914_gel2_HD10.png|thumb|350 px|right|Gel 100914-2]] | [[Image:20100914_gel2_HD10.png|thumb|350 px|right|Gel 100914-2]] | ||
| Line 192: | Line 193: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| - | = 15/09/2010 = | + | ===15/09/2010=== |
[[Image:20100915_gel1_HD2010.png|thumb|350px|right| Gel 100915-1]] | [[Image:20100915_gel1_HD2010.png|thumb|350px|right| Gel 100915-1]] | ||
<br /> | <br /> | ||
| Line 247: | Line 248: | ||
Each PCR was performed in two replicates. The result of the PCRs was analyzed on a 1 % agarose gel (100915-1), run for 1 h @ 100 V. The PCR nr. 2 was subsequently PCR purified by applying a Qiagen PCR purification KIT and used for the cloning | Each PCR was performed in two replicates. The result of the PCRs was analyzed on a 1 % agarose gel (100915-1), run for 1 h @ 100 V. The PCR nr. 2 was subsequently PCR purified by applying a Qiagen PCR purification KIT and used for the cloning | ||
| - | = 16/09/2010 = | + | ===16/09/2010=== |
<br /> | <br /> | ||
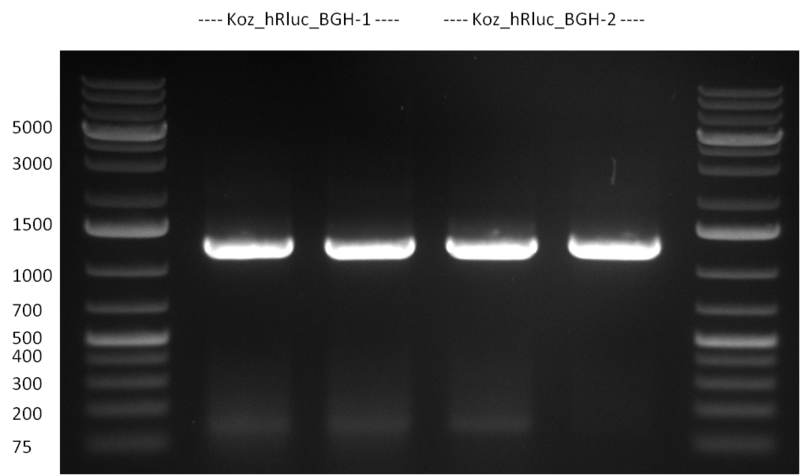
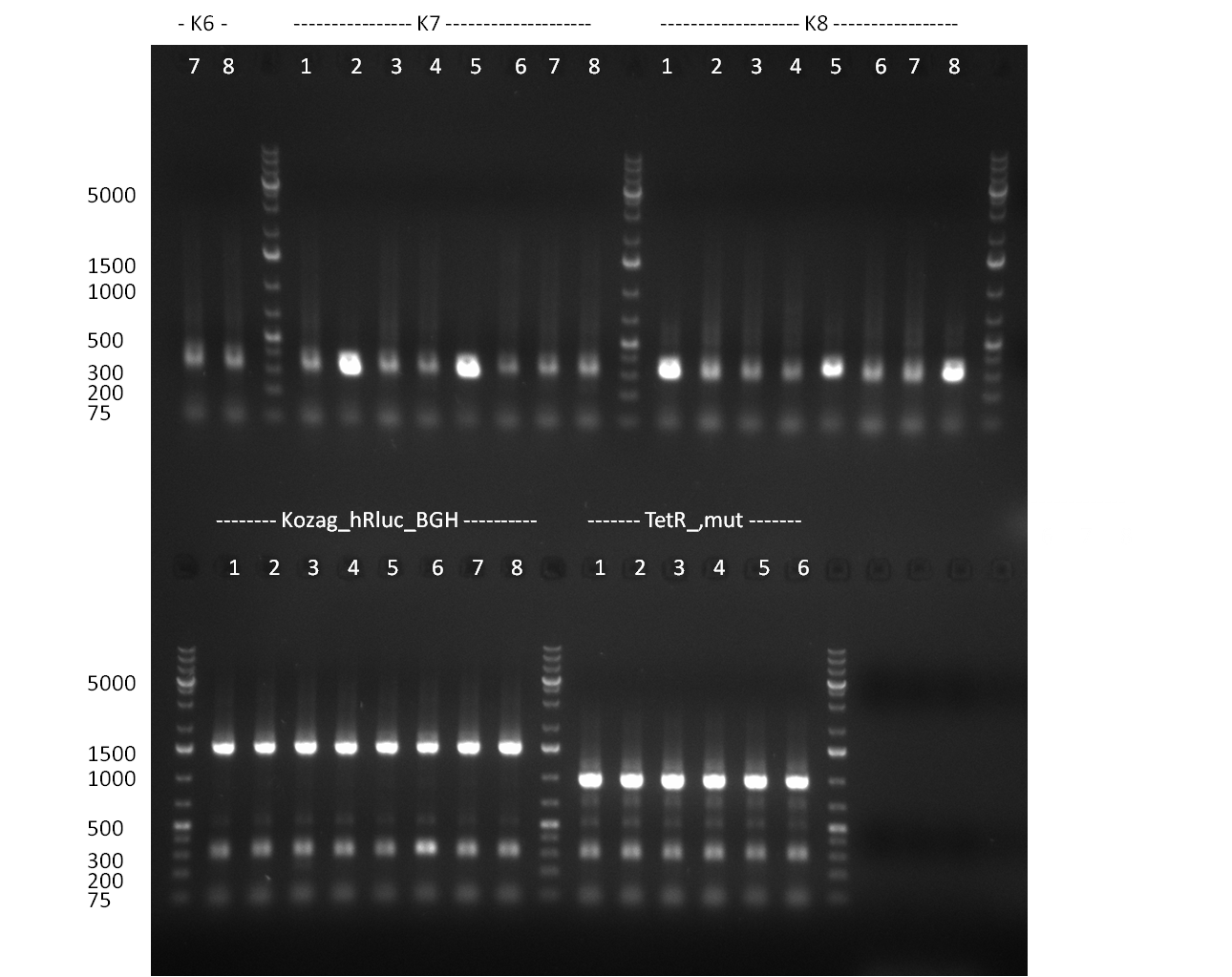
* Cloning of Kozag_hRluc_BGH fragment into pSB1C3 and Constructs R24-R31 | * Cloning of Kozag_hRluc_BGH fragment into pSB1C3 and Constructs R24-R31 | ||
| Line 266: | Line 267: | ||
| - | ==17/09/2010== | + | ===17/09/2010=== |
* colony PCR, mini-prep and digestion of promising psiCHECK or TetR constructs containing binding site | * colony PCR, mini-prep and digestion of promising psiCHECK or TetR constructs containing binding site | ||
** control plates with horrible number of re-ligations | ** control plates with horrible number of re-ligations | ||
| Line 277: | Line 278: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| - | = 19/09/2010 = | + | ===19/09/2010=== |
<br /> | <br /> | ||
* Miniprep of the previous days inocculated miniprep cultures | * Miniprep of the previous days inocculated miniprep cultures | ||
| Line 288: | Line 289: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
| - | = 21/09/2010 = | + | ===21/09/2010=== |
<br /> | <br /> | ||
| - | = 22/09/2010 = | + | ===22/09/2010=== |
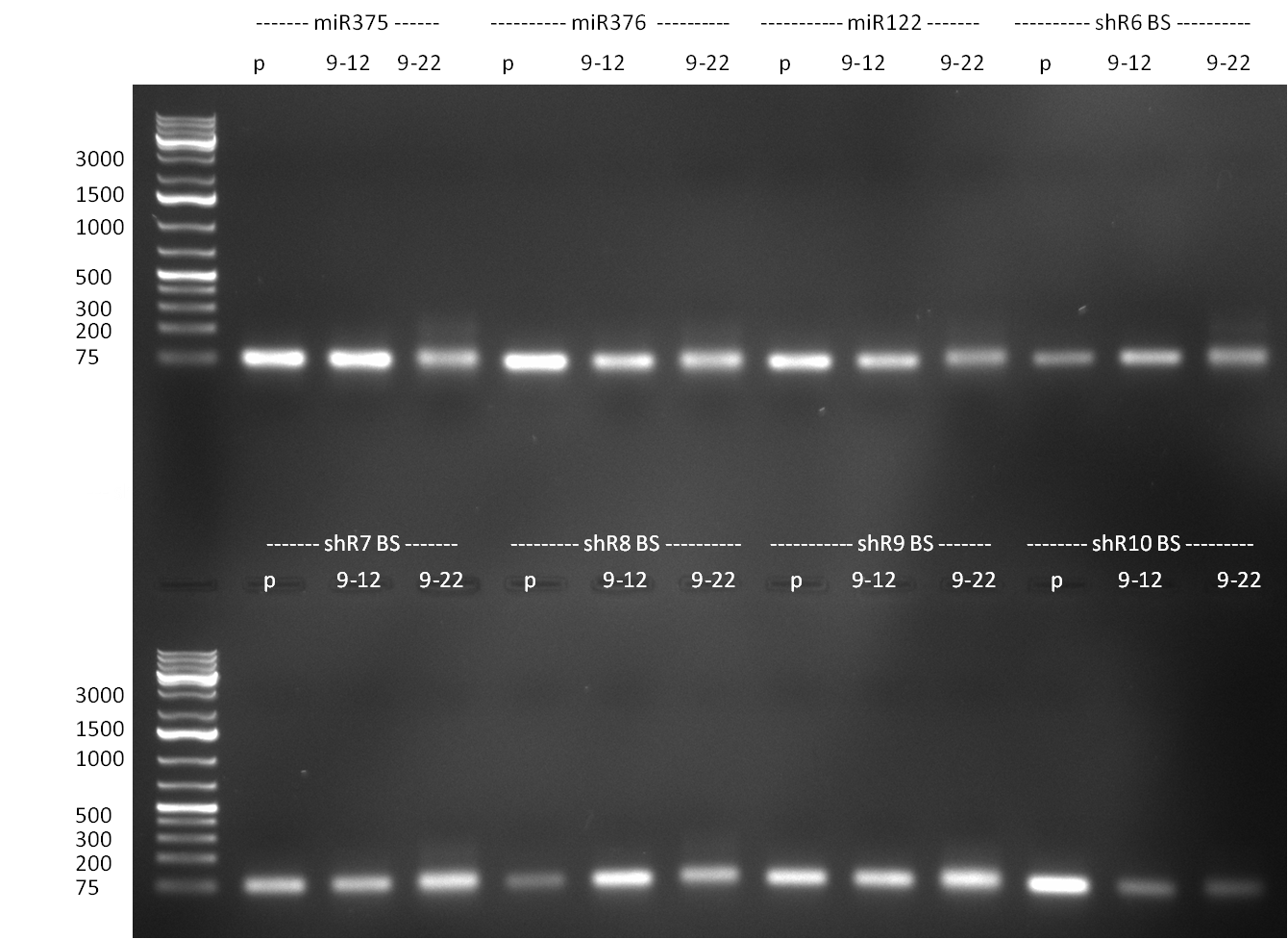
* digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] with EcoRI and PstI in EcoRI Buffer (NEB) and ligation into digested {{Part|pSB1C3}}, transformation, growing on selective agar plates (chloramphenicol), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | * digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] with EcoRI and PstI in EcoRI Buffer (NEB) and ligation into digested {{Part|pSB1C3}}, transformation, growing on selective agar plates (chloramphenicol), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ||
| - | = 23/09/2010 = | + | ===23/09/2010=== |
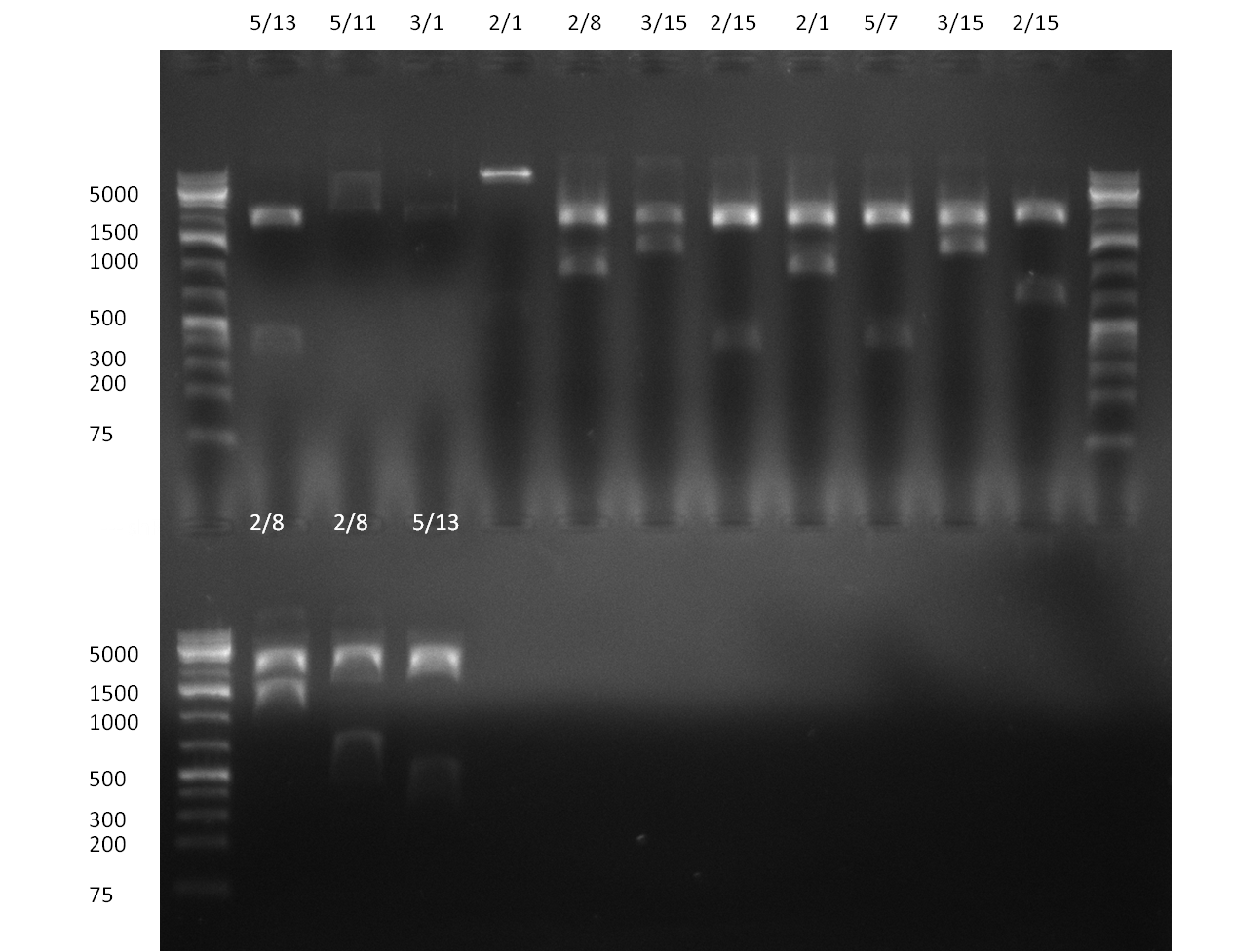
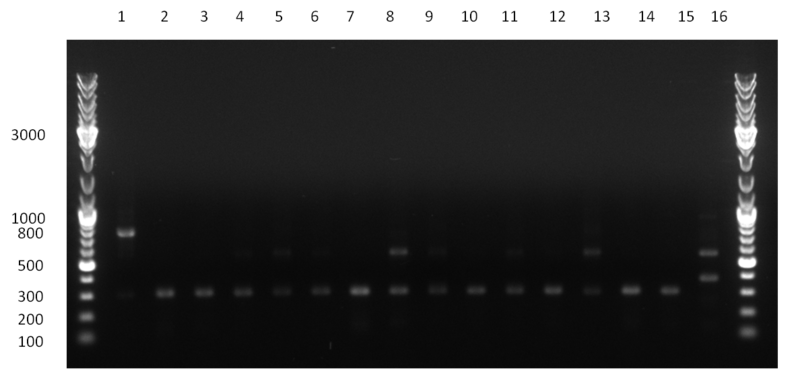
[[Image:20100923 LoLa colonypcr.png | thumb | 350px | right | Gel 100923-1. Analytical gel with following lanes: 1) till 8) colony PCR products with nice bands as expected at <nowiki>~</nowiki> 800 bp especially at lane 1, 4, 6 and 8, lane 9) 1kb Plus Ladder (Invitrogen)]] | [[Image:20100923 LoLa colonypcr.png | thumb | 350px | right | Gel 100923-1. Analytical gel with following lanes: 1) till 8) colony PCR products with nice bands as expected at <nowiki>~</nowiki> 800 bp especially at lane 1, 4, 6 and 8, lane 9) 1kb Plus Ladder (Invitrogen)]] | ||
* minipreps (using Quiagen Kit) and [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of each 8 colonies of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] in {{Part|pSB1C3}} (digested, ligated, transformed and plated on [[Igem2010/Main/synthetic_miR_Kit/September#22/09/2010 | previous day]]) | * minipreps (using Quiagen Kit) and [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of each 8 colonies of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] in {{Part|pSB1C3}} (digested, ligated, transformed and plated on [[Igem2010/Main/synthetic_miR_Kit/September#22/09/2010 | previous day]]) | ||
| Line 300: | Line 301: | ||
** ligation into digested {{Part|pSB1A3}}, [https://2010.igem.org/Transformation transformation], growing on selective agar plates (ampicillin), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ** ligation into digested {{Part|pSB1A3}}, [https://2010.igem.org/Transformation transformation], growing on selective agar plates (ampicillin), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ||
| - | = 24/09/2010 = | + | ===24/09/2010=== |
* "Something is rotten in the state of Genemark..." [[User:Lorenz|Lorenz]] 18:00, 24 September 2010 (UTC) | * "Something is rotten in the state of Genemark..." [[User:Lorenz|Lorenz]] 18:00, 24 September 2010 (UTC) | ||
| Line 312: | Line 313: | ||
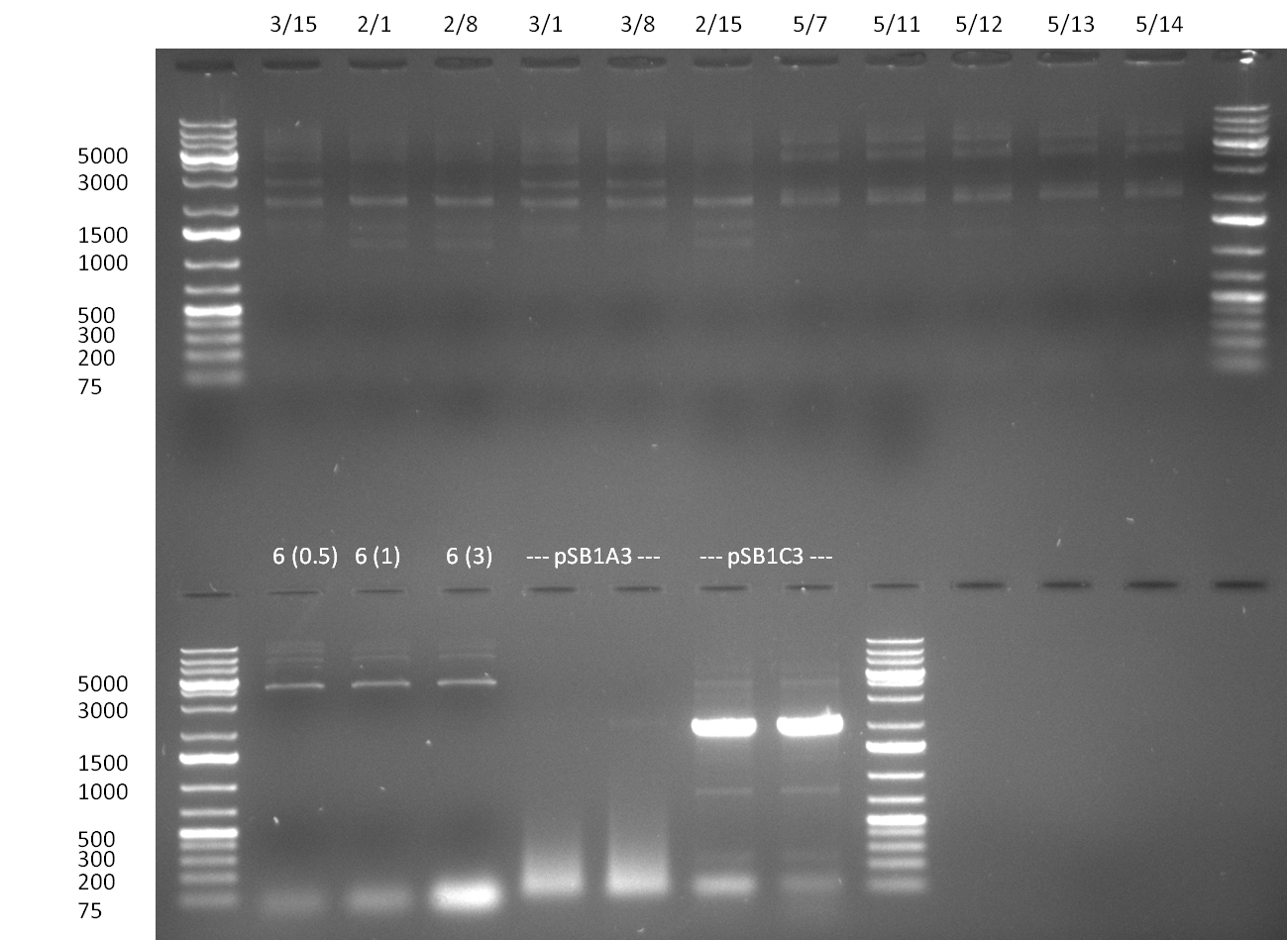
* ligation of R56C + R8 + backbone A | * ligation of R56C + R8 + backbone A | ||
<br> | <br> | ||
| - | = 25/09/2010 = | + | ===25/09/2010=== |
* PCR amplification of standardised pcDNA5 p55 and p1 with primer D47/48 and D55/69 | * PCR amplification of standardised pcDNA5 p55 and p1 with primer D47/48 and D55/69 | ||
| - | =26/09/2010= | + | ===26/09/2010=== |
[[Image:20100926 digestion check.png | thumb | 350px | right | Gel 100926-1. Analytical gel with following lanes: 1) R33 E/N (from 23<sup>rd</sup> September) 2) R23_1 S/P 3) R23_6 4) R33C 5) R23_4 6) R23_8 7) 1kb Plus Ladder (Invitrogen) 8) R33C E/N 9) R23_4 S/P 9) R23C_8 S/P]] | [[Image:20100926 digestion check.png | thumb | 350px | right | Gel 100926-1. Analytical gel with following lanes: 1) R33 E/N (from 23<sup>rd</sup> September) 2) R23_1 S/P 3) R23_6 4) R33C 5) R23_4 6) R23_8 7) 1kb Plus Ladder (Invitrogen) 8) R33C E/N 9) R23_4 S/P 9) R23C_8 S/P]] | ||
[[Image:Rb100927_pcrp55andp1.jpg|thumb|350px|left| Gel 100926-2. PCR results of p55 and p1: p1 with primer 47/48 (lane 1), p1 with primer 55/70 (lane 2), p55 with primer 47/48 (lane3), p55 with primer 55/70]] | [[Image:Rb100927_pcrp55andp1.jpg|thumb|350px|left| Gel 100926-2. PCR results of p55 and p1: p1 with primer 47/48 (lane 1), p1 with primer 55/70 (lane 2), p55 with primer 47/48 (lane3), p55 with primer 55/70]] | ||
| Line 335: | Line 336: | ||
<br><br><br><br> | <br><br><br><br> | ||
| - | = 27/09/2010 = | + | ===27/09/2010=== |
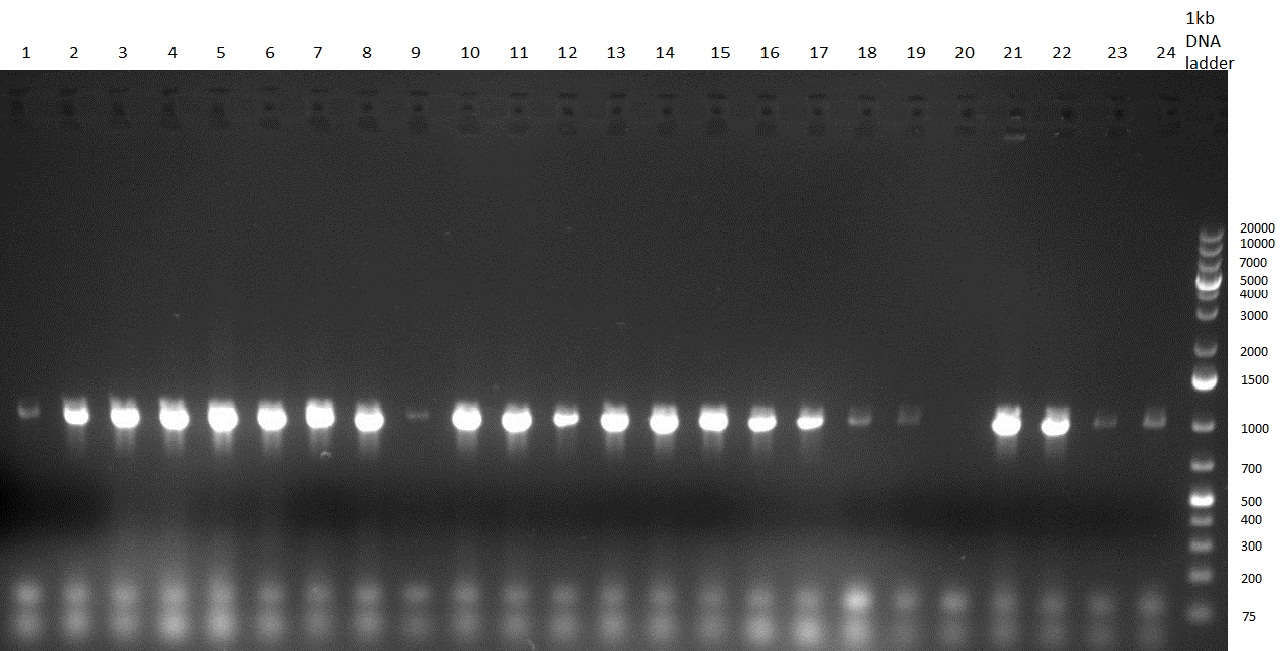
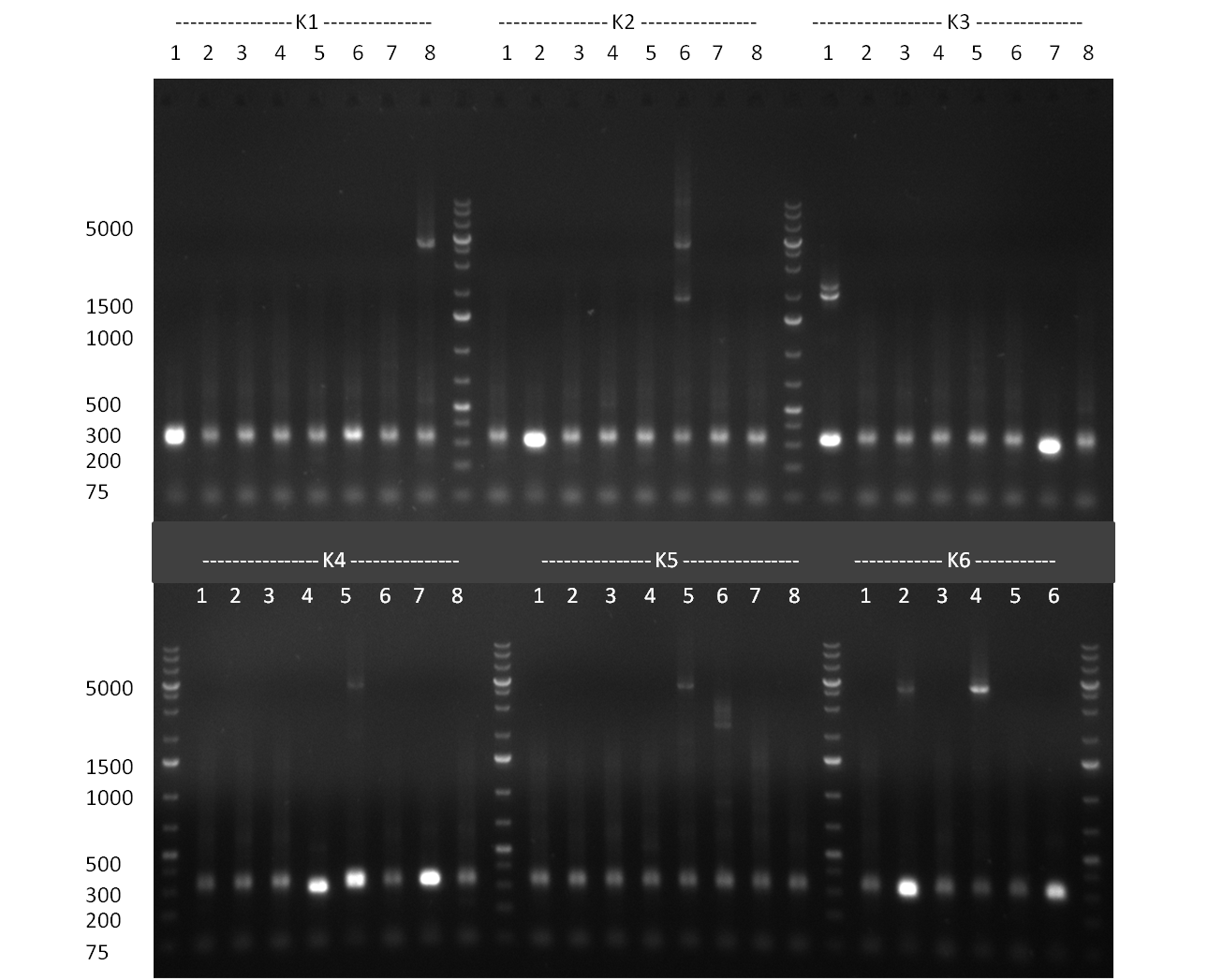
[[Image:Rb100927_colonypcrK14and7aa.jpg|thumb|350 px| right|Gel 100927-1: colony PCR on K1 (lane 1-8), K4 (lane 9-16) and K8 (lane 17-24)]] | [[Image:Rb100927_colonypcrK14and7aa.jpg|thumb|350 px| right|Gel 100927-1: colony PCR on K1 (lane 1-8), K4 (lane 9-16) and K8 (lane 17-24)]] | ||
[[Image:Rb100927_colonypcrK14and7ai.jpg|thumb|350 px|right|Gel 100927-2: colony PCR K1 (lane 1&2), K4 (3&4) und K7 (5&6), K3 digested with BamHI (7), K3 digested NheI and SpeI (8), R33 in pSB1A3 digested with EcoRI and PstI (9), R33 digested with NheI and SpeI (10)]] | [[Image:Rb100927_colonypcrK14and7ai.jpg|thumb|350 px|right|Gel 100927-2: colony PCR K1 (lane 1&2), K4 (3&4) und K7 (5&6), K3 digested with BamHI (7), K3 digested NheI and SpeI (8), R33 in pSB1A3 digested with EcoRI and PstI (9), R33 digested with NheI and SpeI (10)]] | ||
| Line 360: | Line 361: | ||
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
| - | = 28/09/2010 = | + | ===28/09/2010=== |
[[Image:Rb100928_K3.jpg|thumb|350px|middle| Gel 100928-1. Colony PCR of K3 construct]] | [[Image:Rb100928_K3.jpg|thumb|350px|middle| Gel 100928-1. Colony PCR of K3 construct]] | ||
[[Image:Rb100928_K3andtestdigestK147.jpg|thumb|350px|right| Gel 100928-2. Test digest of with PstI and EcoRI (lane 1-6), test digest with BamHI (lane 7-12), K1 (lane 1&2), K4 (lane 3&4), K7 (lane 5&6)]] | [[Image:Rb100928_K3andtestdigestK147.jpg|thumb|350px|right| Gel 100928-2. Test digest of with PstI and EcoRI (lane 1-6), test digest with BamHI (lane 7-12), K1 (lane 1&2), K4 (lane 3&4), K7 (lane 5&6)]] | ||
| Line 386: | Line 387: | ||
<br><br><br><br> | <br><br><br><br> | ||
| - | = 29/09/2010 = | + | ===29/09/2010=== |
[[Image:Rb100928testpe_bk1k3.jpg|thumb|350px|left|Gel 100929-1 first gel: double digestion of K1 and K3 with PstI and EcoRI: K1 (lane 1-4), K3 (lane 5-8), second gel: digestion of K1 and K3 with BamHI: K1 (lane 1-4), K3 (lane 5-8)]] | [[Image:Rb100928testpe_bk1k3.jpg|thumb|350px|left|Gel 100929-1 first gel: double digestion of K1 and K3 with PstI and EcoRI: K1 (lane 1-4), K3 (lane 5-8), second gel: digestion of K1 and K3 with BamHI: K1 (lane 1-4), K3 (lane 5-8)]] | ||
[[Image:Rb100929k1k3ahdigest.jpg|thumb|350px|right|Gel 1000929-2 digestion of K1 and K3 with HindIII and AflII: K1 (lane 1&2), K3(lane 3-12)]] | [[Image:Rb100929k1k3ahdigest.jpg|thumb|350px|right|Gel 1000929-2 digestion of K1 and K3 with HindIII and AflII: K1 (lane 1&2), K3(lane 3-12)]] | ||
| Line 426: | Line 427: | ||
<br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
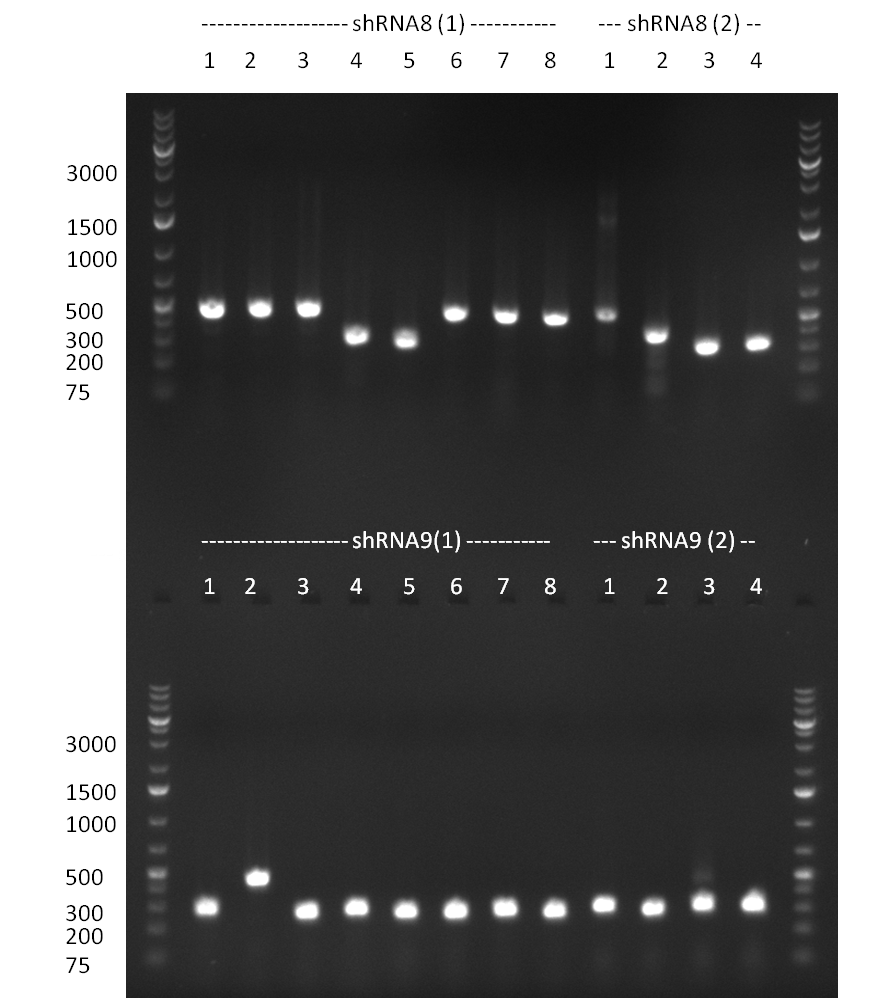
| - | = 30/09/2010 = | + | ===30/09/2010=== |
[[Image:20100930_gel1.jpg|thumb|350px|right|Gel 1000903-2 colony PCR on shRNA10 cloned into K3 and K4]] | [[Image:20100930_gel1.jpg|thumb|350px|right|Gel 1000903-2 colony PCR on shRNA10 cloned into K3 and K4]] | ||
[[Image:20100930_gel2.jpg|thumb|350px|right|Gel 1000930-3 colony PCR on shRNA10 into K3 and K4]] | [[Image:20100930_gel2.jpg|thumb|350px|right|Gel 1000930-3 colony PCR on shRNA10 into K3 and K4]] | ||
Revision as of 14:43, 25 October 2010

|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"