Team:TU Munich/Lab
From 2010.igem.org
FPraetorius (Talk | contribs) (→Chronological Lab Book) |
FPraetorius (Talk | contribs) (→Chronological Lab Book) |
||
| Line 1,271: | Line 1,271: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | {{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| - | + | ===31.05.2010=== | |
| + | * Digestions | ||
| + | **15N-1 (BBa_I719005, 19.5 ng/uL) with S/P | ||
| + | **HisSig/TrpSig (6.5/10 ng/uL) with X/P | ||
| + | ::2 h 37 °C | ||
| + | :heat inactivation of insert-digestions | ||
| + | |||
| + | *Gel: 1% Agarose in 1x TAE | ||
| + | :1 h 25 min, 115 V | ||
| + | :stained with SybrGold, 40 min | ||
| + | :[[File:100531.png]] | ||
| + | |||
| + | :Band at ~2100b cut und purified using the [[protocols#ZYMO RESEARCH Gel DNA Recovery Kit |zymo kit]] | ||
| + | |||
| + | * [[protocols#Ligation|ligation]] | ||
| + | ** HisSig (4 uL of digest) with purified plasmid (with BBa_I719005) | ||
| + | ** TrpSig (2 uL of digest) with purified plasmid -=- | ||
| + | ::reason: concentration of His Sig before digest was 1/2 of TrpSig | ||
| + | |||
| + | * [[protocols#Transformation|transformation]] | ||
| + | : of DH5a with Ligation batches, HisSig1-3, HisSig3-1, TrpSig DL4-1, TrpSig 4-3 | ||
| + | ===01.06.2010=== | ||
| + | *'''[[Protocols#PCR|Colony PCR]]''' | ||
| + | |||
| + | :of Ligations transformed into DH5a yesterday | ||
| + | :4 Colonies of each Ligation | ||
| + | |||
| + | **T7-HisSig | ||
| + | **T7-TrpSig | ||
| + | |||
| + | <br> | ||
| + | |||
| + | *'''Gel: 3% broad range Agarose in 1xTBE''' | ||
| + | |||
| + | :1.5 h 140 V | ||
| + | :stained with SybrGold | ||
| + | |||
| + | [[Image:100601.png]] | ||
| + | |||
| + | :calculation for the expected size of the fragments | ||
| + | |||
| + | :{| cellspacing="1" cellpadding="1" border="1" width="200" | ||
| + | |- | ||
| + | | part | ||
| + | | size (bp) | ||
| + | |- | ||
| + | | HisSig | ||
| + | | 32 | ||
| + | |- | ||
| + | | TrpSig | ||
| + | | 34 | ||
| + | |- | ||
| + | | T7 promoter | ||
| + | | 23 | ||
| + | |- | ||
| + | | prefix | ||
| + | | 20 | ||
| + | |- | ||
| + | | suffix | ||
| + | | 21 | ||
| + | |- | ||
| + | | X/S scar | ||
| + | | 6 | ||
| + | |} | ||
| + | |||
| + | :in PCR we get additional bp due to the primers - +9 at pre/suffix=+18 bp<br> | ||
| + | |||
| + | :overall size of the fragments expected to come out of the PCR: T7_HisSig: 120 bp, T7_TrpSig: 122 bp | ||
| + | |||
| + | *'''ON cultures''' | ||
| + | |||
| + | :5 ml cultures of pSB1K3_R0011_HisSig_B0014 (1_3 & 3_1) and pSB1K3_R0011_TrpSig_B0014 (DL4_1 & DL4_3) | ||
| + | :1 ml cultures of each colony monitored in Colony PCR | ||
| + | |||
| + | ===02.06.2010=== | ||
| + | *Miniprep of yesterdays cultures using Zymokit, elution by nuclease-free water | ||
| + | *Concentration determination | ||
| + | *analytic digestion | ||
| + | *results on gel: | ||
| + | |||
| + | <br> | ||
| + | |||
| + | *Sequencing <br> | ||
| + | |||
| + | JobNr. Barcode Last change Date/Time Last message / Files 6549287 AE2739 02.06.2010 / 13:51:12 HisSig 1-3-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | 6549288 AE2738 02.06.2010 / 13:51:12 HisSig 3-1-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | 6549289 AE2737 02.06.2010 / 13:51:12 TrpSig DL4-1-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | 6549290 AE2736 02.06.2010 / 13:51:12 TrpSig DL4-3-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | 6549291 AE2735 02.06.2010 / 13:51:12 HisTerm-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | 6549292 AE2734 02.06.2010 / 13:51:12 TrpTerm-forward G1004 | ||
| + | |||
| + | We just received your order. Many thanks. | ||
| + | |||
| + | *Gel 3% broad range agarose in 1x TBE | ||
| + | : [[File:100602_beschr.png]] | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 1,278: | Line 1,388: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | {{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| - | + | ===07.06.2010=== | |
| + | *'''Sequenbcing results from GATC''' | ||
| + | **HisSig DL1-3 is ok | ||
| + | **HisSig 3-1 is ok | ||
| + | **TrpSig DL4-1 is ok | ||
| + | **TrpSig DL4-3 is ok | ||
| + | **TrpTerm + HisTerm bad runs... --> new sequencing order with Primer 100 bp upstream (within GFP) | ||
| + | |||
| + | :Files can be found stored in our [[Zugangsdaten für GATC|GATC account]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | *'''Sequencing@GATC:''' both Term-constructs with primer pGFP-FP provided by GATC | ||
| + | |||
| + | <br> | ||
| + | |||
| + | *'''[[Protocols#Restriction|Restrictions]] ''' | ||
| + | **psB1A10_TrpTerm/HisTerm with Nsi1, Aat2 | ||
| + | **pSB1K3_R0011_HisSig/TrpSig_B0014 with Pst1, Aat2 | ||
| + | **T7 bb with Spe1, Pst1 | ||
| + | **PCRProducts: HisSig/TrpSig with Pst1, Xba1, 2 h @37°C | ||
| + | |||
| + | :all plasmid digests done''' sequential''' as enzymes do not have 100% activity in the same buffer, each reaction 1.5 h@37°C | ||
| + | :psB1A10_TrpTerm/HisTerm and T7 bb dephosphorylated the last 30 min <br> | ||
| + | |||
| + | *'''[[Protocols#Ligation|Ligation]] ''' <br> | ||
| + | *'''[[Protocols#Transformation|Transformation]] ''' | ||
| + | |||
| + | <br> liquid culture (10 ml) of pSB1K3_R0011_HisSig/TrpSig_B0014 | ||
| + | ===08.06.2010=== | ||
| + | *'''Sequencing results from GATC''': His/TrpTerm with pGFP-FP primer | ||
| + | **HisTerm worked | ||
| + | **TrpTerm worked | ||
| + | : checked with blast2seq | ||
| + | |||
| + | *'''[[Protocols#PCR|Colony PCR]]''' | ||
| + | : 8 colonies from each plate of T7His, T7Trp, MonsterHis, MonsterTrp as ligations resulted in many colonies | ||
| + | |||
| + | *'''3% Agarose Gel in 1xTBE''' | ||
| + | : for all colony PCR reactions | ||
| + | [[File:100608_beschriftet.png]] | ||
| + | [[File:100608-2_beschriftet.png]] | ||
| + | |||
| + | <br> Interpretation/Info: The R0011_Sig_B0014 construct was cut with PstI, but not ligated into a PstI site but instead into the NsiI site --> even if sticky end are compatible, the bases in the 3` direction are different --> primer lags 7 bp compared to standard procedure --> we didn´t expect to find the signal construct by colony PCR --> control digestion tomorrow | ||
| + | : T7-Signal constructs seem to have worked, expected size was 23 bp (T7)+ 32 (HisSig)/34 (TrpSig) bp + 30+29 (PCRPre+Suf)=114/116 bp | ||
| + | *over night cultures | ||
| + | |||
| + | ===09.06.2010=== | ||
| + | * Miniprep of 4 Monster_His, 4 Monster_Trp, 3 T7_His and 3 T7_Trp cultures | ||
| + | * Analytical digestion of plasmids mentioned above | ||
| + | * gel of Monster_Plasmid digestion | ||
| + | [[File:100609_Monster_inverse.jpg]] | ||
| + | <br> | ||
| + | gel didn´t work at all --> even after > 2h, bands were not separated correctly, even the 1kb ladder was "stacked" in the gel-pockets, the 100 bp ladder should show EQUAL distances between the lines [http://www.neb.com/nebecomm/productfiles/778/images/N3231_fig1_v1_000034.gif see here], it looks like the gel was "more dense" at the pockets---> no idea what happened --> repeat Monster-digestion tomorrow? | ||
| + | <br> | ||
| + | * gel of T7_Plasmid digestion | ||
| + | [[File:100609_T7_sig_invers.jpg]] | ||
| + | <br> | ||
| + | T7_Trp E + S digestion 107 bp and T7_His 105 bp --> worked for all picked colonies. (regard that there is an excess of plasmid DNA-basepairs of factor >30 --> thats why the inserts are much weaker than the plasmid signals. | ||
| + | <br> | ||
| + | occured trouble: | ||
| + | ** Ladders and loading dye´s empty --> i used those of eike, BUT: eikes 1 kb ladder is different --> compare [http://www.neb.com/nebecomm/products/productN3272.asp here] and his loading dye was much more diluted, even if there was also 6x Sac GLP written on it -> i hope this won´t cause any trouble | ||
| + | |||
| + | ===10.06.2010=== | ||
| + | *'''ordered''' | ||
| + | :Promega E.coli S30 in vitro transcription/translation kit | ||
| + | :Spe1, Aat2 from NEB, 500 U each | ||
| + | <br> | ||
| + | * '''[[Protocols#Restriction|analytical Digestion]] | ||
| + | :of Ligation colonies from MonsterHis/trp 1-3 and T7His/Trp 1,2,5/1,2,3 | ||
| + | :2h digestion | ||
| + | :: Monster: 6 uL DNA template with Aat2/Spe1 in Buffer 4/Bsa | ||
| + | :: T7-Signal: 6 uL DNA template with E/P in Buffer 3 | ||
| + | <br> | ||
| + | * '''Agarose Gels''' | ||
| + | :used standards: lmw, 2-log [[Protocols#standards|click here]] | ||
| + | : Gel1: 1% Agarose in 1xTBE for Digestions of Monsterplasmid | ||
| + | :: run in big chamber @ 200 V for 1 h 20 min | ||
| + | :[[File:100610_t7sig.png]] | ||
| + | :Gel2: 3% Agarose (broad range) in 1xTBE for Digestions of T7-Signal | ||
| + | :: run in small chamber @140 V for 1 h 35 min | ||
| + | :[[File:100610_monster.png]] | ||
| + | <br><br> | ||
| + | :Conclusions: | ||
| + | # all T7-Signal ligations loaded on the gel worked | ||
| + | # monsterplasmid didn't work? bands at 800 bp, 900 bp, 1.3 kbp, 2.2 kbp, 3 kbp, we SHOULD expect to see our Insert, wich is Prefix+R0011_Signal_B0014_small Suffix, which should run around 300-400 bp... | ||
| + | |||
| + | ===11.06.2010=== | ||
| + | *'''Gel''': large 1% Agarose in TAE. Load: The rest of N/A cut Messplasmids from [[07.06.2010]]. Run @220 V for 3.5 h | ||
| + | : fragments expected are 5087 and 176. original size of plasmid is 5263. This is a Try to differ between 5087 and 5263 bp | ||
| + | :[[File:100611.png]] | ||
| + | : Band @ 5000 bp of Trp_Term purified, obviously digestion was 100%. Bad oint is that HisTerm includes an Nsi1 cleavage site... | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 1,285: | Line 1,487: | ||
{{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | {{:Team:TU Munich/Templates/ToggleBoxStart2}} {{:Team:TU_Munich/Templates/RedBox | text=Cloning }} {{:Team:TU_Munich/Templates/BlueBox | text=Measurements }} {{:Team:TU_Munich/Templates/GreenBox | text=Measurements }} {{:Team:TU_Munich/Templates/YellowBox | text=Measurements }} | ||
{{:Team:TU Munich/Templates/ToggleBoxStart3}} | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| - | + | ===14.06.2010=== | |
| + | *'''[[Protocols#Ligation|Ligation]]''' | ||
| + | : 10 uL pSB1A10_TrpTerm Aat2/Nsi1 0.5 ng/uL | ||
| + | : 1 uL R0011_TrpSig_B0014 Aat2/Psb1 11 ng/uL | ||
| + | : 2 uL T4 ligase buffer | ||
| + | : 1 uL T4 Ligase | ||
| + | : 6 uL H2O | ||
| + | ::10 min RT | ||
| + | <br> | ||
| + | *'''Transformation''' of DH5a with | ||
| + | : Ligation | ||
| + | : T7His#1 | ||
| + | : T7Trp#1 | ||
| + | <br> | ||
| + | *'''over night cultures''' of | ||
| + | : pSB1K3_R0011_TrpSig_B0014 | ||
| + | : pSB1K3_R0011_HisSig_B0014 | ||
| + | : pSB1A10_TrpTerm | ||
| + | : pSB1A10_HisTerm | ||
| + | |||
| + | ===15.06.2010=== | ||
| + | *Over night cultures | ||
| + | *Aliquots of the Promega in vitro expressions kit from e. coli s30 extract: | ||
| + | : 40 uL with aa mix including all aa. | ||
| + | |||
| + | ===16.06.2010=== | ||
| + | ====Fluoresence measurements using in vitro kit==== | ||
| + | * in vitro kit sample | ||
| + | * adding psBA1A10 Trp_Term --> constant over time, no significant changes compared to kit alone --> high efficiency of AraC | ||
| + | * adding L-(+)Arabinose (final concentration 2%) --> after approx. 10 min significant GFP production --> measuring for xxx min --> RFP is slightly increased (to proof if correlated to GFP peak --> crossdetection) | ||
| + | * adding psB1K3 R0011_TrpSig_B0014 | ||
| + | ====Cell culture==== | ||
| + | 5 ml culture for | ||
| + | * psBA1A10 Trp_Term/HisTerm | ||
| + | * psB1K3 R0011_TrpSig/Hissig_B0014 | ||
| + | ===17.06.2010=== | ||
| + | ==== Cloning ==== | ||
| + | <br> | ||
| + | Digestion of Trp-Sig with E/P and psB1A10 Trp_Term with E/P | ||
| + | * Gel purification of psB1A10 Trp_Term E/P cut | ||
| + | [[File:100617_pSB1A10_EPcut_dunkler.jpg]] | ||
| + | * Heat inactivation of Trp-Sig E/P cut | ||
| + | * Ligation for 10 min @ RT and Transformation in DH5-a cells | ||
| + | |||
| + | ==== in vitro measurements ==== | ||
| + | |||
| + | ===18.06.2010=== | ||
| + | ==== cloning ==== | ||
| + | * Transformation (about 20 colonies) --> picking 5 colonies | ||
| + | * colony PCR | ||
| + | * Gel <br> | ||
| + | 2 % broad range agarose, 1 h 120 V [[File:100618_psb1A10-Trp_sig_colonypcr.jpg]] | ||
| + | Sample 2, 4, 5 shows probably Trp-Signal + Pre/Suffix --> send sample 2 for sequencing! | ||
| + | |||
| + | <br><br> | ||
| + | * control digestion of all 10 picked psB1A10-TrpSig in 1% broad range agarose, > 3 h, 120 V | ||
| + | [[File:100618_psb1A10-Trp_sig_controlverdau.jpg]] | ||
| + | --> digestions worked, but again, no insert can be found, despite gel was at maximum resolution ( 3h 120 V, see LMW) | ||
| + | |||
| + | ==== in vitro measurements ==== | ||
| + | f$%&&§ s%§$! again, nothing worked! despite we saw an increasing "GFP signal" comparable to 16.06.10, taking spectra suggested we DONT see significant GFP-production! We used new water for preparing the samples, cleaned cuvettes with "new water", used other DNA-samples etc. Somehow, it seems like we dont express GFP (we compared Christophs results! One should see a really significant spectrum! <br>Next steps:* Try in vivo measurements, just using psb1A10_xTerm without Signal (thus just measuring plasmid) to proof if kit or measuring plasmid causes this problem! | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
Revision as of 21:27, 24 October 2010
|
|
Experiment DesignWe designed different experimental set-ups with varying complexity to test RNA signal/switch pairs based on our concept. In vivo MeasurementsIn vivo measurements have the highest complexity compared to any other experimental set-up. Our system has to deal with several circumstances a cellular environment comes with, such as interaction with other RNAs, degradation by RNases or unspecific interactions. Nevertheless, the measurements are essential, as our switches should finally work inside cells to fulfill our vision of an intracellular logic network.
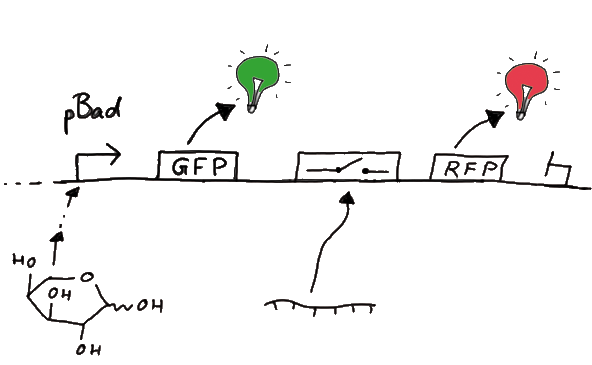
DesignFor the measurements in vivo we decided to use an expression cassette consisting of Green Fluorescent Protein (GFP) coding sequence upstream of the switch and another fluorescent protein coding sequence downstream of it. Both protein coding sequence carry the same ribosome binding site, therefore, the GFP fluorescence can be used as internal control in measurements. Since the spectra should not overlap and to avoid FRET as well as an pure overlap of the spectra, we settled on the usage of red fluorescent protein variants, namely mRFP1 in the first try. While the GFP fluorescence is used to normalize the measurements, the RFP fluorescence is used to detect termination/antitermination.
Upon binding of the signal, the stem loop of the switch would resolve leading to red fluorescence. The GFP fluorescence as internal control carries the advantage that errors in the measurement set can be detected easily. Lack of arabinose or promoter insensitivity can be recognized as well as problems with the fluorescence measurement itself. Plus, we have a way to normalize our measurements and compare different preparations in relation to each other.
Construction and CloningOur measuring plasmid is based on the BioBrick pSB1A10, A1, distribution 2010. Unfortunately after two months of cloning we had to recognize that the plasmid in use did not work (see also Biobrick validation--> link). So after the first unsuccessful attempts we decided to reclone the system, substituing RFP to mCherry, a dsRED derivative with a spectrum in the far red, and adding arabinose inducible promoters in front of both fluorescent proteins.
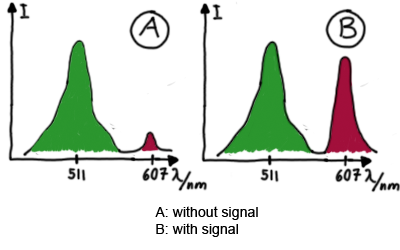
MeasurementFor switch evaluation, IPTG was added to the cells after about two hours after arabinose induction (baseline). ??? Stimmt das?? A rise of RFP/mCherry emission should be visible in case of a working switch.
When measuring the termination of our BioBricks and the antitermination by their corresponding signal-RNA, we should be able to observe an increasing RFP emission compared to the GFP emission upon induced signal-RNA production in the cells/in the kit:
With these measurements, it should also be possible to observe differences in efficiency of termination as well as antitermination between our designed switches. In vitro TranslationIn vitro measurements with E. coli lysate make the fluorescence signals independent of cell growth and physical or biological factors, e.g. cell density or growth stadium.
DesignIn this assay we used the same constructs as engineered for the in vivo studies. MeasurementsWe used the cell-free E. coli S30 extract system for circular DNA provided by promega[1], which is prepared by modifications of the Method Zubay et al.[2] described. The characterization of the kit can be obtained from the [http://partsregistry.org/Cell-free_chassis/Commercial_E._coli_S30 Parts Registry]. In vitro TranscriptionAn experiment, in which we detect In vitro transcription, offers an elegant way for a fast and easy prove of principle, since our switch is RNA-based and the whole mechanism takes place on trancriptional level. Most side effects occuring in a complex environment given in a cell or a cell lysate do not arise here.
If measureable effects with our basic concept can be seen in vitro we can use the so gained data to optimize the system in vivo. Since we are working on a totally new principle of trancriptional control, we used this approach for easy variation of different variables like the length of the core unit and the switch to signal ratio.
To study the switches on the transcriptional level gives the advantage, that we would have less interferences and possible artefacts. Also, we are not sure how cellular mechanisms like degradation of RNases or interacting factors as well as molecular crowding influence our systems.
T7 RNA polymeraseThe T7 RNA polymerase is known for satisfying RNA yields together with easy handling. In our approach we had PCR amplified, double stranded switches with an malachitegreen binding aptamer following after the switch (133 bp, see section below) and a single stranded signal with about 30 bp length.
For in vitro expression the T7 RNA Polymerase requires a double stranded promotor region at the beginning of the DNA template but is otherwise capable of handling single stranded DNA, so a sense strain corresponding to the T7 promoter region was added. Transcription is more effective with double stranded DNA as template. Since we ordered the signal sequences we tested we chose the cheaper way in the beginning by using single stranded signals with corresponding sense T7 pieces and switched to double stranded constructs after narrowing down the most promising switch/signal pairs.
E. coli RNA polymeraseIn comparison to the T7 RNA Polymerase the E. coli RNA Polymerase requires slightly more sophisticated proceedings when it comes to the design of switches and handling of the enzyme. The biggest in our case was to store it properly since the only -80°C fridge was in another building. Denaturing Polyacrylamide gel electrophoresisWe also used Polyacrylamide gel electrophoresis (PAGE) for evaluation of termination efficiency of our basic units. Gels containing 15 % acrylamide and 6 M urea were used for separation of 90 (terminated by switch) and 133 bp (continous reading) RNAs.
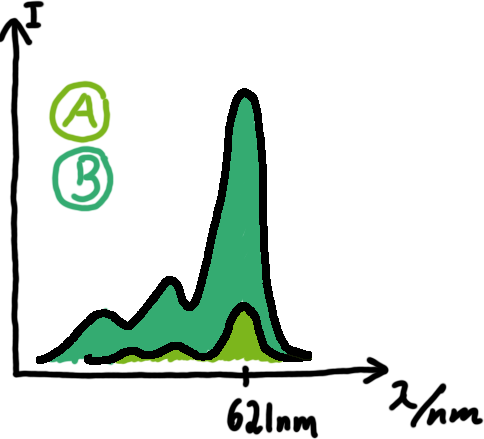
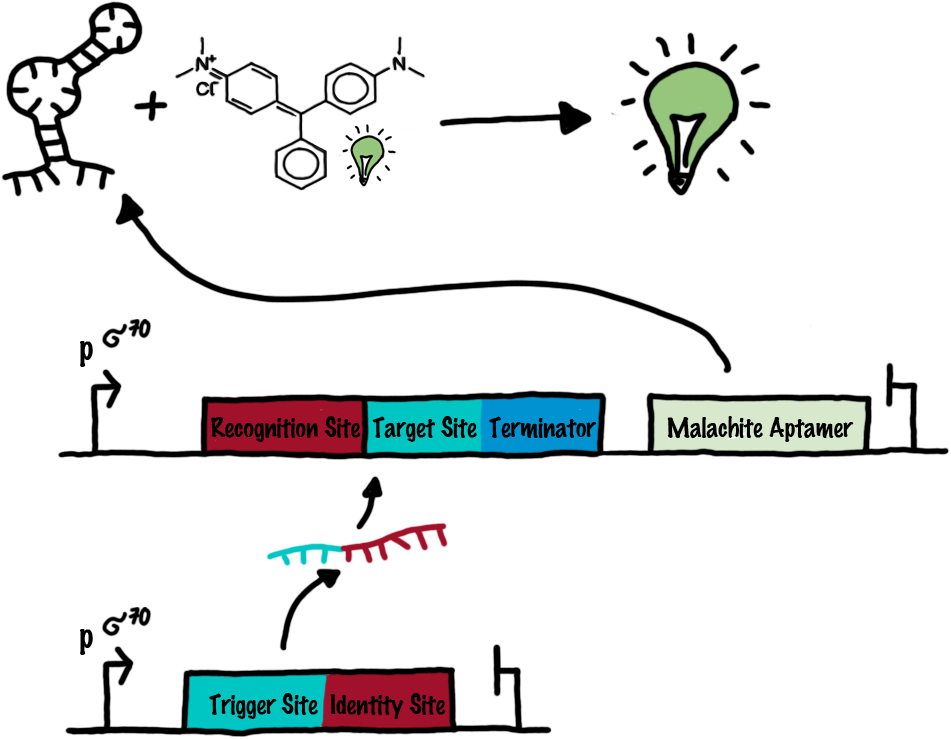
Malachite green assayMalachite-green is a dye with a negligible fluorescence in solution but undergoes a dramatic increase if bound by a RNA -aptamer. Upon binding to the aptamer, the fluorescence of malachite-green increases about 3000 times making it an exceptionel good marker. Since the binding is very specific, transcription in dependence of a signal can be monitored by measuring the fluorescence of malachite-green over time if the aptamer is located behind the switch. Transcription of the aptamer will only take place after anti-termination by a signal. An increase should be visible over time.
OLD: A second possibility to measure parameters of our switches we came up with, was the idea to investigate our system on the transcriptional level only. Therefore, we decided to use malachite green as reporter. Malachite green in a fluorescent dye, whose emission increasing dramaticly (about 3000 times) upon binding of a specific RNA-aptamer. The RNA-aptamer
We made constructs comprising of a sigma(70)-binding promoter followed by a short nonsense sequence, the switches and the aptamer sequence.
References[1] http://www.promega.com/catalog/catalogproducts.aspx?categoryname=productleaf_335&ckt=1 [2] Zubay, G. (1980) Meth. Enzymol. 65, 856–77, Zubay, G. (1973) Ann. Rev. Genet. 7, 267–87. Experimental ResultsIn vivo MeasurementsIn vivo In vitro Measurements
ProtocolsMolecular BiologyPCR
So geht ne PCR CloseDigestion
So geht ne PCR CloseLigation
In vivo Measurement
So haben wir in vivo gemessen CloseIn vitro Translation
So haben wir in vitro gemessen CloseIn vitro Transcription
So haben wir in vivo gemessen CloseLab BookExplanationsIn the following we present an overview regarding our work in the lab. For easier understanding we summarized the work of each week using colored boxes. To get more information about the work and results of a specific week, just click on the according week number. To get a better overview we used the following color code for the boxes:
Chronological Lab Book
Constrcuts for in vivo measurments
08.04.2010Flo & Philipp PCR
09.04.2010Philipp & Flo Gel of PCR products from 08.04.2010
Improved Constructs... Testing
15.04.2010Philipp & Flo
16.04.2010Philipp & Flo
--> worked for R0011, not for B0014
* approximated from the amount used in the digestion beforeClose
Measurements
19.04.2010
* approximated from the amount used in the digestion before 20.04.2010
21.04.2010
--> worked!!!!!
22.04.2010
23.04.2010
Cloning Measurements Measurements Measurements
26.04.2010
27.04.2010
Many colonies with pSB1K3-B0014, not one with pSB1K3-Sig-B0014
28.04.2010
29.04.2010
30.04.2010PCR R0011-HisSig and R0011-TrpSig --> 13 ng/µl x 20 µl Close
Cloning Measurements Measurements Measurements
04.05.2010
--> each in 600 µl LB+Carbenicillin (=Ampicillin) @37°C 05.05.2010
File:100505beschriftet.png
06.05.2010
07.05.2010
Cloning Measurements Measurements Measurements
10.05.2010
Prefix: 20 bp; Suffix: 21 bp; X-S-scar: 6 bp --> it looks as if R0011 is ligated to B0014, which makes the whole construct wothless. The R0011-HisSig control looks more like R0011 alone as well. 11.05.2010
12.05.2010
14.05.2010
Cloning Measurements Measurements Measurements
17.05.2010
18.05.2010
Samples: LMW|R0011|HisSig|TrpSig|HisSig E/S-Dig|TrpSig E/S-Dig|B0014|LMW2|Colony PCR His1|His2|Trp1|Trp2|control|HisTerm|TrpTerm|HisTerm E/P-Dig|TrpTerm E/P-Dig
Important Mistake! See below Gel!
File:100518beschriftet.png
(*R0011 and B0014 look normal
--> are all of our sequences just wrong??? What are we going to do? Order everything new?) 19.05.2010
20.05.2010
21.05.2010
Cloning Measurements Measurements Measurements
25.05.2010
26.05.2010
27.05.2010
28.05.2010
Cloning Measurements Measurements Measurements
31.05.2010
01.06.2010
02.06.2010
JobNr. Barcode Last change Date/Time Last message / Files 6549287 AE2739 02.06.2010 / 13:51:12 HisSig 1-3-forward G1004 We just received your order. Many thanks. 6549288 AE2738 02.06.2010 / 13:51:12 HisSig 3-1-forward G1004 We just received your order. Many thanks. 6549289 AE2737 02.06.2010 / 13:51:12 TrpSig DL4-1-forward G1004 We just received your order. Many thanks. 6549290 AE2736 02.06.2010 / 13:51:12 TrpSig DL4-3-forward G1004 We just received your order. Many thanks. 6549291 AE2735 02.06.2010 / 13:51:12 HisTerm-forward G1004 We just received your order. Many thanks. 6549292 AE2734 02.06.2010 / 13:51:12 TrpTerm-forward G1004 We just received your order. Many thanks.
Cloning Measurements Measurements Measurements
07.06.2010
08.06.2010
File:100608 beschriftet.png File:100608-2 beschriftet.png
09.06.2010
File:100609 Monster inverse.jpg
File:100609 T7 sig invers.jpg
10.06.2010
11.06.2010
Cloning Measurements Measurements Measurements
14.06.2010
15.06.2010
16.06.2010Fluoresence measurements using in vitro kit
Cell culture5 ml culture for
17.06.2010Cloning
File:100617 pSB1A10 EPcut dunkler.jpg
in vitro measurements18.06.2010cloning
2 % broad range agarose, 1 h 120 V File:100618 psb1A10-Trp sig colonypcr.jpg Sample 2, 4, 5 shows probably Trp-Signal + Pre/Suffix --> send sample 2 for sequencing!
File:100618 psb1A10-Trp sig controlverdau.jpg --> digestions worked, but again, no insert can be found, despite gel was at maximum resolution ( 3h 120 V, see LMW) in vitro measurementsf$%&&§ s%§$! again, nothing worked! despite we saw an increasing "GFP signal" comparable to 16.06.10, taking spectra suggested we DONT see significant GFP-production! We used new water for preparing the samples, cleaned cuvettes with "new water", used other DNA-samples etc. Somehow, it seems like we dont express GFP (we compared Christophs results! One should see a really significant spectrum!
Cloning Measurements Measurements Measurements
This is what we did in week xxx. Close
Cloning Measurements Measurements Measurements
This is what we did in week xxx. Close
Cloning Measurements Measurements Measurements
This is what we did in week xxx. Close
Cloning Measurements Measurements Measurements
This is what we did in week xxx. Close
Cloning Measurements Measurements Measurements
This is what we did in week xxx. Close |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"