Team:Imperial College London/Lab Diaries/Vectors team
From 2010.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
{{:Team:Imperial_College_London/Templates/LabDiariesHeader}} | {{:Team:Imperial_College_London/Templates/LabDiariesHeader}} | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | + | ||
| - | | | + | <html> |
| - | | | + | <head> |
| - | |} | + | <style type="text/css"> |
| - | + | #content {background-image: url(http://openwetware.org/images/2/29/Kkbgd2.JPG)} | |
| - | + | </style> | |
| - | | | + | </head> |
| - | | | + | <body> |
| - | | | + | </body> |
| + | </html> | ||
| + | [[Image:Kkbgd3.JPG]] | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
| + | <html> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | ||
| + | <title>Imperial iGEM 2010</title> | ||
| + | <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.4.2/jquery.min.js"></script> | ||
| + | <script type="text/javascript"> | ||
| + | $(document).ready(function(){ | ||
| + | $("ul[id|=poplist]").hide(); | ||
| + | |||
| + | $("p[id|=popshow]").click(function() { | ||
| + | $("ul[id|=poplist]").slideToggle("slow"); | ||
| + | return false; | ||
| + | }); | ||
| + | |||
| + | |||
| + | }); | ||
| + | </script> | ||
| + | <style type="text/css"> | ||
| + | #popcon | ||
| + | { | ||
| + | width: 360px; | ||
| + | } | ||
| + | |||
| + | #popcon ul | ||
| + | { | ||
| + | margin-left: 0px; | ||
| + | padding-left: 0px; | ||
| + | list-style: none; | ||
| + | font-family: Arial, Helvetica, sans-serif; | ||
| + | } | ||
| + | |||
| + | #popshow | ||
| + | { | ||
| + | width: 360px; | ||
| + | margin-left: 0px; | ||
| + | padding-left: 0px; | ||
| + | list-style: none; | ||
| + | font-family: Arial, Helvetica, sans-serif; | ||
| + | display: block; | ||
| + | color: #fff; | ||
| + | text-decoration: none; | ||
| + | text-align: center; | ||
| + | background-color: #CCCCCC; | ||
| + | padding: 3px; | ||
| + | border-bottom: 1px solid #fff; | ||
| + | } | ||
| + | |||
| + | #popcon a:link, #popcon a:visited | ||
| + | { | ||
| + | display: block; | ||
| + | color: #fff; | ||
| + | text-decoration: none; | ||
| + | text-align: center; | ||
| + | background-color: #CCCCCC; | ||
| + | padding: 3px; | ||
| + | //border-bottom: 1px solid #FFF; | ||
| + | } | ||
| + | |||
| + | #popcon a:hover, #popcon a:active | ||
| + | { | ||
| + | background-color: #FFCCFF | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | <body> | ||
| + | <p id="popshow">Follow our progress: Click me!</p> | ||
| + | <div id="popcon"> | ||
| + | <ul id="poplist"> | ||
| + | <li><a href="#Objectives" class="popt">Objectives</a></li> | ||
| + | <li><a href="#Assembly_of_Vectors" class="popt">Assembly of Vectors</a></li> | ||
| + | <li><a href="#Schedule" class="popt">Schedule</a></li> | ||
| + | <li><a href="#Lab_Notes" class="popt">Lab Notes</a></li> | ||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
| + | ==Objectives== | ||
| + | |||
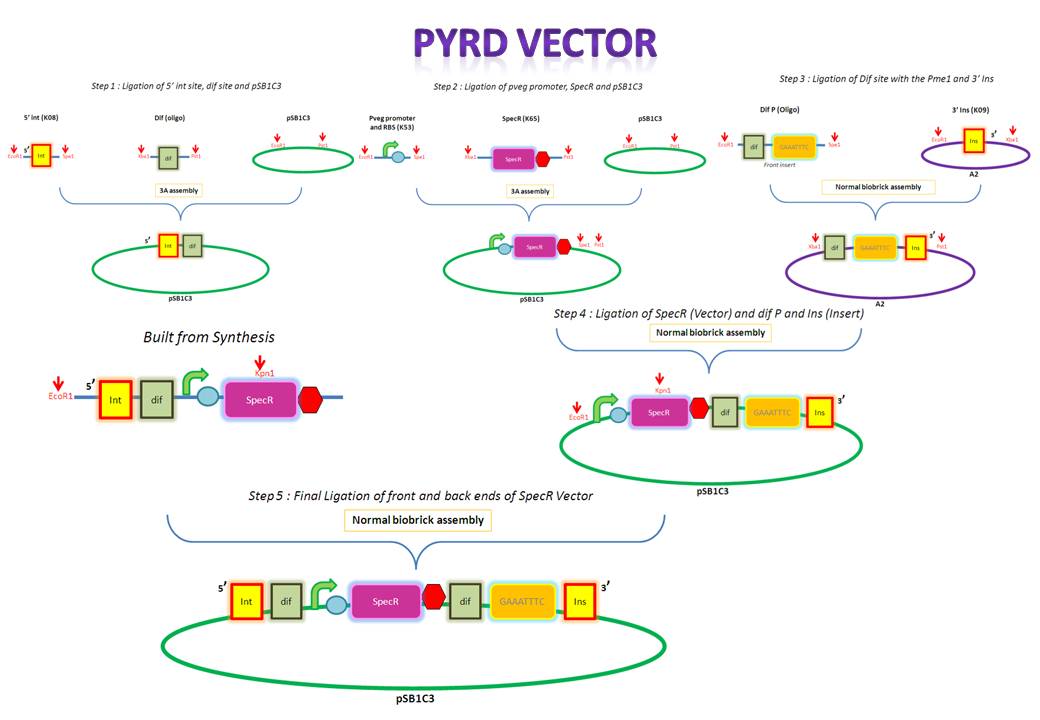
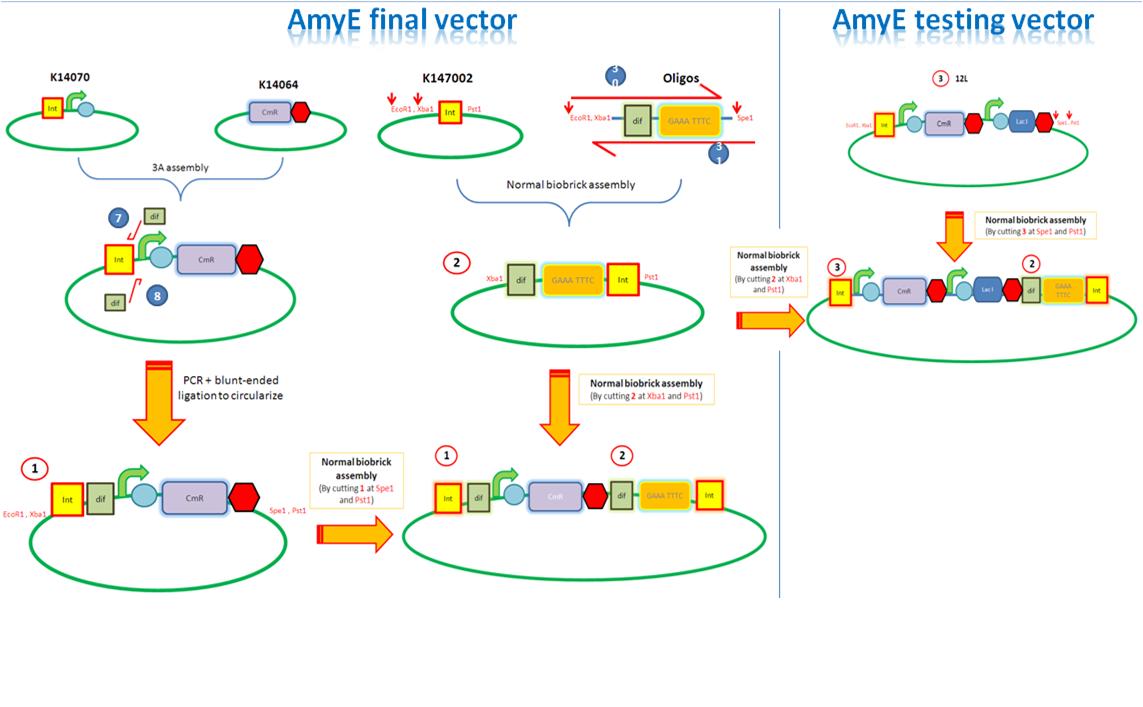
| + | We are assembling the AmyE and PyrD vectors in order to transform B. Subtilis with our parts. Once completed, these vectors will be reusable and can then be used to introduce any relevant piece of DNA directly into B.subtilis genome. The vectors will be used both for the final assembly and for testing constructs. | ||
| + | |||
| + | *[[IGEM:Imperial/2010/PyrD_Vector | Assembly of the<font color = #6B3FA0> '''PyrD vector''' </font>]] | ||
| + | *[[IGEM:Imperial/2010/AmyE_Vector | Assembly of the<font color = #CA2C92> '''AmyE vector'''</font>]] | ||
| + | |||
| + | ==Assembly of Vectors== | ||
| + | |||
| + | [[Image:IC_PyrD.JPG |750px|thumb|center|alt=A|Overveiw of PyrD assembly]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:IC_AmyE.JPG |750px|thumb|center|alt=A|Overveiw of AmyE assembly]] | ||
Revision as of 15:23, 23 October 2010
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
 "
"