Team:Imperial College London/Modelling/Output/Detailed Description
From 2010.igem.org
(Editing 1st para) |
(Many changes mainly in abandoned attempts) |
||
| Line 11: | Line 11: | ||
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Output Amplification Model | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Output Amplification Model | ||
|- | |- | ||
| - | |< | + | | |
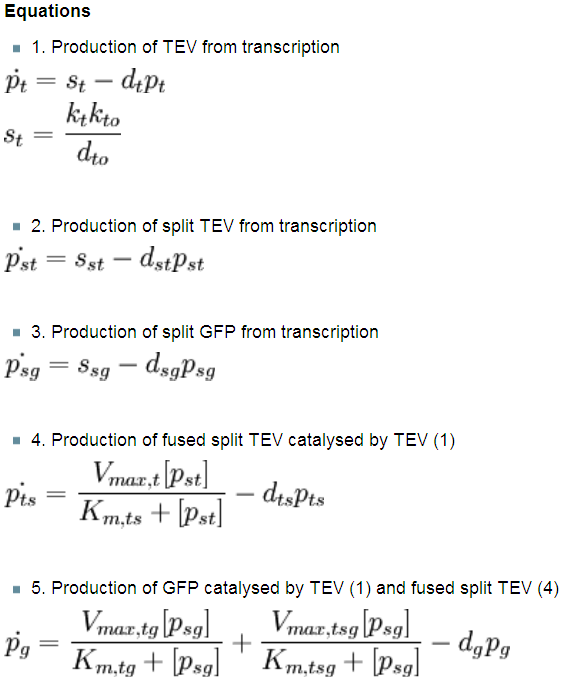
| - | < | + | The following page presents the details of the models that have been developed. Firstly, assumptions that have been exploited are explained. Then every model is presented separately as each of them has slightly different elements of the system and the interactions between them. However, there are only 3 fundamental biochemical processes that will be analysed: |
| + | <ol> | ||
| + | <li>Transcription</li> | ||
| + | <li>Translation</li> | ||
| + | <li>Enzymatic reactions</li> | ||
| + | </ol> | ||
| - | During a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. Click on the button below to | + | <html><h2>1. Law of Mass Action</h2></html> |
| + | |||
| + | During a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. | ||
| + | |||
| + | A few of Michaelis-Menten assumptions were not met by our system: | ||
| + | *'''Vmax is proportional to the overall concentration of the enzyme.''' | ||
| + | Since we are continuously producing enzyme, Vmax will change. Therefore the conservation <html>E<sub>0</sub> = E + E<sub>S</sub></html> does not hold for our system. | ||
| + | *'''Substrate >> Enzyme''' | ||
| + | We are producing both substrate and enzyme, so we have approximately the same amount of substrate and enzyme. | ||
| + | *'''Enzyme affinity to the substrate has to be high.''' | ||
| + | |||
| + | |||
| + | Click on the button below, in order to get to know more on Michaelis-Menten kinetics based models. | ||
<div class="accordionButton">Abandoned Initial Attempts</div> | <div class="accordionButton">Abandoned Initial Attempts</div> | ||
<div class="accordionContent"> | <div class="accordionContent"> | ||
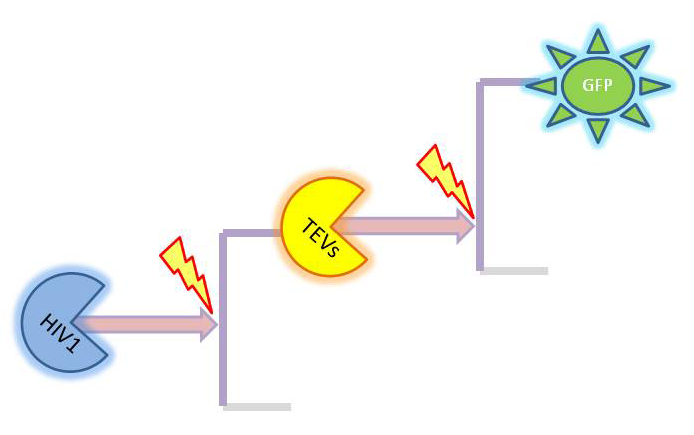
| - | < | + | <b>Elements of the system</b><br/> |
| - | + | Depending on which amplification it there is several species appearing. The full list is: | |
| + | *GFP (on diagrams shown <i>green</i>) which was out original choice for output later on changed to dioxygenase acting on catechol. It was supposed to be split and attached to coiled coils. It would get activated by TEV protease. | ||
| + | *TEV protease (shown <i>orange</i> on diagrams)and split TEV (TEVs is shown <i>yellow</i>). Split TEV would be inactive form mounted on the coiled coils to be activated by HIV1 or another active TEV. | ||
| + | *HIV1 protease (shown <i>blue</i> on diagrams) which would be produced active upon receiving the activation signal in case of 2 amplification step outputs. | ||
| - | < | + | <b>Simple models</b><br/> |
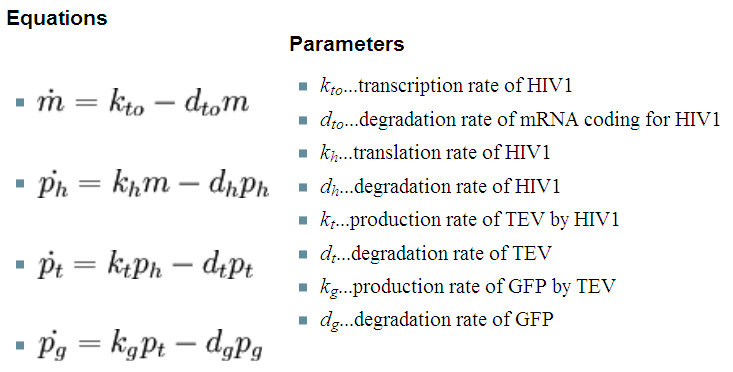
| - | [[Image: | + | Our first models only were accounting for the protein and mRNA production: |
| + | |||
| + | {|style="width:825px;background:#eeeeee;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<div ALIGN=Center> | ||
| + | {| style="width:270px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Initial_1.png|268px]] | ||
| + | |- | ||
| + | |2-step amplifier with distinct protease at each level. | ||
| + | |} | ||
| + | </div> | ||
| + | | <div ALIGN=Center> | ||
| + | {| style="width:545px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Initial_1_eqn.png|543px]] | ||
| + | |} | ||
| + | </div> | ||
| + | |- | ||
| + | | | ||
| + | |Where: m - is the concentration of mRNA, p - is the protein concentration (subscript indicates which protein) | ||
| + | |- | ||
| + | | <div ALIGN=Center> | ||
| + | {| style="width:270px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
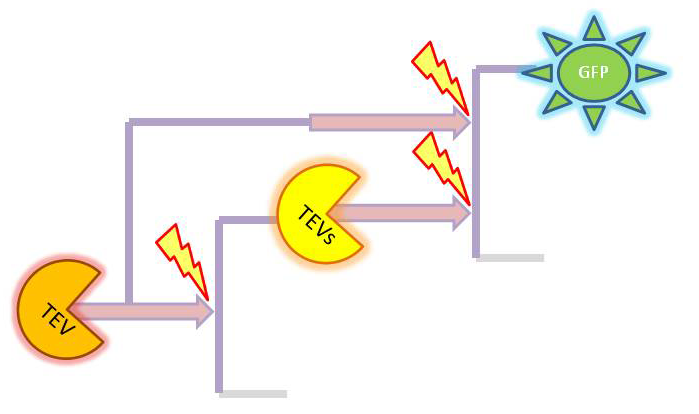
| + | |[[Image:Initial_2.png|268px]] | ||
| + | |- | ||
| + | |2-step amplifier with TEV protease implemented at both amplification levels. | ||
| + | |} | ||
| + | </div> | ||
| + | |<div ALIGN=Center> | ||
| + | {| style="width:545px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Initial_2_eqn.png|543px]] | ||
| + | |} | ||
| + | </div> | ||
| + | |} | ||
| + | |||
| + | <b>Implementation of Michaelis-Menten kinetics</b><br/> | ||
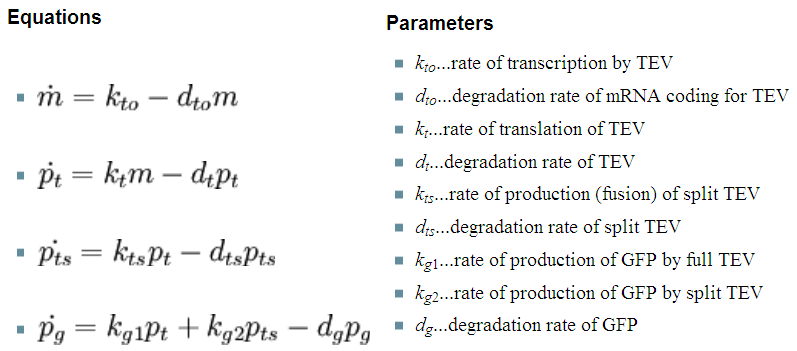
| + | So, far the models were considering only the production and degradation of species. The example presented below includes enzymatic interaction between the species governed by Michaelis-Menten. | ||
| + | {|style="width:825px;background:#eeeeee;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<div ALIGN=Center> | ||
| + | {| style="width:470px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Initial_1.png|468px]] | ||
| + | |- | ||
| + | |2-step amplifier with distinct protease at each level. | ||
| + | |} | ||
| + | </div> | ||
| + | | <div ALIGN=Center> | ||
| + | {| style="width:345px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 1px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Initial_3_eqn.png|343px]] | ||
| + | |} | ||
| + | </div> | ||
| + | |} | ||
<br/><br/> | <br/><br/> | ||
| Line 90: | Line 166: | ||
</html> | </html> | ||
| - | < | + | <b>Conclusion</b><br/> |
We were not able to obtain all the necessary constants. Hence, we decided to make educated guesses about possible relative values between the constants as well as varying them and observing the change in output. | We were not able to obtain all the necessary constants. Hence, we decided to make educated guesses about possible relative values between the constants as well as varying them and observing the change in output. | ||
As the result, we concluded that the amplification happens at each amplification level proposed. The magnitude of amplification varies depending on the constants. There is not much difference between using TEV or HIV1. | As the result, we concluded that the amplification happens at each amplification level proposed. The magnitude of amplification varies depending on the constants. There is not much difference between using TEV or HIV1. | ||
| + | |||
| + | <b>Change of output</b> | ||
| + | |||
| + | During our literature research, we came across a better output, so we abandoned the idea of using GFP as an output. Instead, we are using catechol. An enzyme, dioxygenase, will be acting on the catechol, which will then result in a coloured output. Catechol will be added to the bacteria manually (i.e. the bacteria will not produce catechol). Hence, in our models dioxygenase will be treated as an output as this enzyme is the only activator of catechol in our system. This means that the change of catechol into its colourful form is dependent on the dioxygenase concentration. | ||
<h2>References</h2> | <h2>References</h2> | ||
#Kapust R. et al (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Engineering. [Online] 14(12), 993-1000. Available from: http://peds.oxfordjournals.org/cgi/reprint/14/12/993 [Accessed 28th July 2010] | #Kapust R. et al (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Engineering. [Online] 14(12), 993-1000. Available from: http://peds.oxfordjournals.org/cgi/reprint/14/12/993 [Accessed 28th July 2010] | ||
| - | </div><br/><br/> | + | </div> |
| - | + | <br/><br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | As we could not use the Michaelis-Menten simplification to model enzymatic reactions in our system, we will had to solve the problem from first principle. It meant referring to more general set of assumptions called Law of Mass Action. This allowed us to model our enzymatic reactions without making assumptions about amounts of particular species as long as the amounts are bigger than single molecule level. This resulted in bigger number of partial differential equations as there was one per each species instead of 1 per reaction. | ||
| - | <html>< | + | <html><h2>2. Model preA: Simple production of dioxygenase</h2></html> |
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| Line 125: | Line 196: | ||
This model includes transcription and translation of the dioxygenase. It does not involve any amplification steps. It is our control model against which we will be comparing the results of other models. | This model includes transcription and translation of the dioxygenase. It does not involve any amplification steps. It is our control model against which we will be comparing the results of other models. | ||
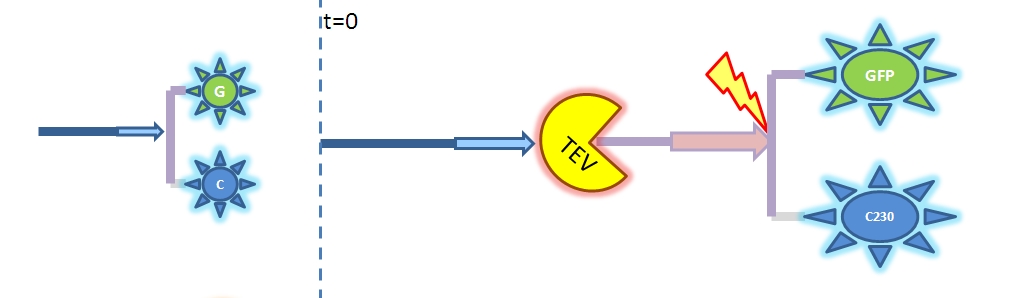
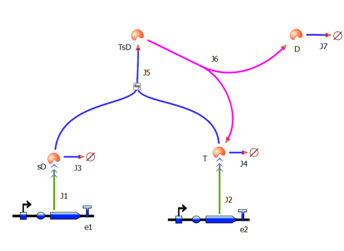
| - | <html>< | + | <html><h2>3. Model A: Activation of Dioxygenase by TEV enzyme</h2></html> |
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| Line 162: | Line 233: | ||
</div> | </div> | ||
| - | <html>< | + | <html><h2>4. Model B: Activation of Dioxygenase by TEV or activated split TEV enzyme</h2></html> |
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| Line 183: | Line 254: | ||
| - | <html>< | + | <html><h2>5. Model C: Further improvements</h2></html> |
This model has not been implemented because of the conclusions that we reached from Models A and B. | This model has not been implemented because of the conclusions that we reached from Models A and B. | ||
Revision as of 00:30, 22 October 2010
| Temporary sub-menu: Objectives; Detailed Description; Parameters & Constants; Results & Conclusion;Download MatLab Files; |
| Output Amplification Model | ||||||||||||||||||||||||||||
|
The following page presents the details of the models that have been developed. Firstly, assumptions that have been exploited are explained. Then every model is presented separately as each of them has slightly different elements of the system and the interactions between them. However, there are only 3 fundamental biochemical processes that will be analysed:
1. Law of Mass ActionDuring a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. A few of Michaelis-Menten assumptions were not met by our system:
Since we are continuously producing enzyme, Vmax will change. Therefore the conservation E0 = E + ES does not hold for our system.
We are producing both substrate and enzyme, so we have approximately the same amount of substrate and enzyme.
Abandoned Initial Attempts
Elements of the system
Simple models
Implementation of Michaelis-Menten kinetics
Conclusion We were not able to obtain all the necessary constants. Hence, we decided to make educated guesses about possible relative values between the constants as well as varying them and observing the change in output. As the result, we concluded that the amplification happens at each amplification level proposed. The magnitude of amplification varies depending on the constants. There is not much difference between using TEV or HIV1. Change of output During our literature research, we came across a better output, so we abandoned the idea of using GFP as an output. Instead, we are using catechol. An enzyme, dioxygenase, will be acting on the catechol, which will then result in a coloured output. Catechol will be added to the bacteria manually (i.e. the bacteria will not produce catechol). Hence, in our models dioxygenase will be treated as an output as this enzyme is the only activator of catechol in our system. This means that the change of catechol into its colourful form is dependent on the dioxygenase concentration. References
As we could not use the Michaelis-Menten simplification to model enzymatic reactions in our system, we will had to solve the problem from first principle. It meant referring to more general set of assumptions called Law of Mass Action. This allowed us to model our enzymatic reactions without making assumptions about amounts of particular species as long as the amounts are bigger than single molecule level. This resulted in bigger number of partial differential equations as there was one per each species instead of 1 per reaction. 2. Model preA: Simple production of dioxygenaseThis model includes transcription and translation of the dioxygenase. It does not involve any amplification steps. It is our control model against which we will be comparing the results of other models. 3. Model A: Activation of Dioxygenase by TEV enzyme
 This is a simple enzymatic reaction, where TEV is the enzyme, Dioxygenase the product and split Dioxygenase the substrate. Choosing k1, k2, k3 as reaction constants, the reaction can be rewritten in these four sub-equations:
 These four equations were implemented in Matlab, using a built-in function (ode45) which solves ordinary differential equations. Implementation in TinkerCell Another approach to model the amplification module would be to implement it in a program such as TinkerCell (or CellDesigner). This would be useful to check whether the Matlab model works. 4. Model B: Activation of Dioxygenase by TEV or activated split TEV enzymeThis version includes the following features:
5. Model C: Further improvementsThis model has not been implemented because of the conclusions that we reached from Models A and B. It would include the following features:
|
 "
"