Team:Imperial College London/Modelling
From 2010.igem.org
(Difference between revisions)
(CHanging diagrams) |
(Updating amplification diagrams) |
||
| Line 68: | Line 68: | ||
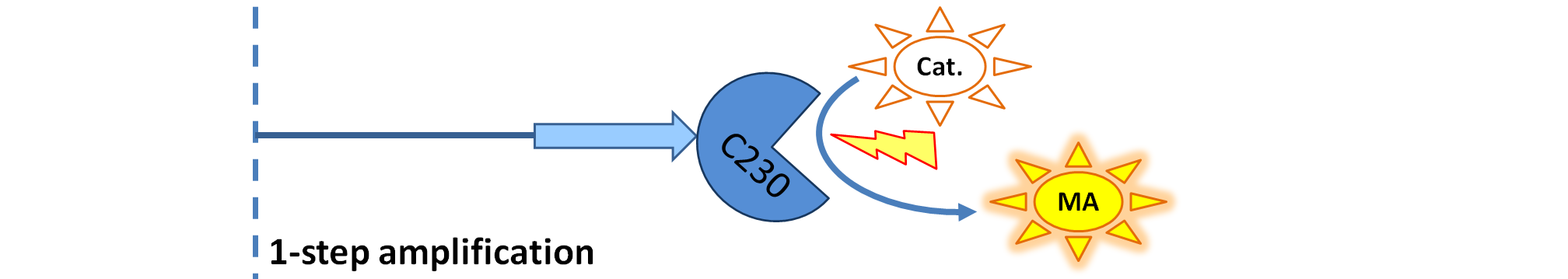
<ol><li>Dioxygenase (<i>blue on the diagrams below</i>) is an enzyme that acts on catechol to produce a yellow output. In most of our models dioxygenase was treated as an output because it was found that active dioxygenase acting on catechol produces the coloured output within a split second.</li> | <ol><li>Dioxygenase (<i>blue on the diagrams below</i>) is an enzyme that acts on catechol to produce a yellow output. In most of our models dioxygenase was treated as an output because it was found that active dioxygenase acting on catechol produces the coloured output within a split second.</li> | ||
<li>GFP-Dioxygenase fusion protein (<i>GFP is shown green on the diagrams</i>). Dioxygenase joined by the linker to GFP was assumed to be inactive.</li> | <li>GFP-Dioxygenase fusion protein (<i>GFP is shown green on the diagrams</i>). Dioxygenase joined by the linker to GFP was assumed to be inactive.</li> | ||
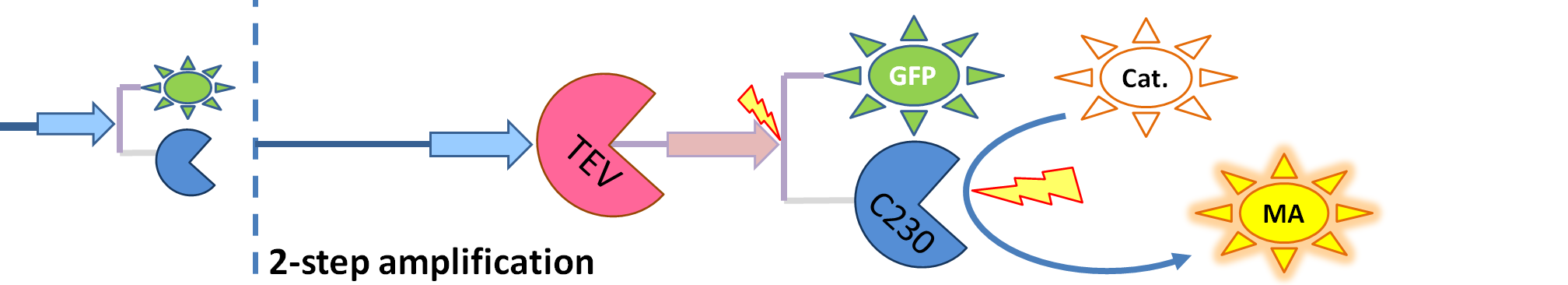
| - | <li>TEV protease (<i> | + | <li>TEV protease (<i>pink on the diagrams below</i>) has the ability to cleave the GFP-Dioxygenase fusion protein, hence, it activates dioxygenase</li> |
| - | <li>Split TEV protease (<i> | + | <li>Split TEV protease (<i>purple on the diagrams below</i>) is an inactive split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.</li> |
</ol> | </ol> | ||
| Line 75: | Line 75: | ||
{| style="background:#e7e7e7;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5" width=704px; | {| style="background:#e7e7e7;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5" width=704px; | ||
|- | |- | ||
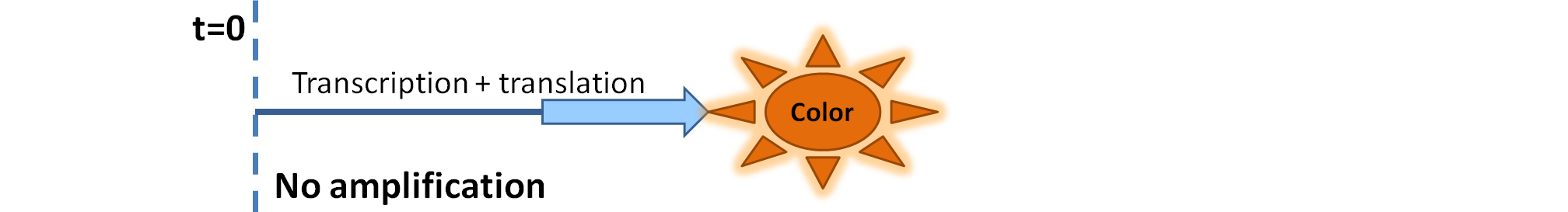
| - | |[[Image: | + | |[[Image:Simple_Production.png|700px]] |
|- | |- | ||
| - | | | + | |Simple production upon activation of arbitrary colour output by transcription and translation indicated by the <i>blue arrow</i>. |
| + | |- | ||
| + | |[[Image:1-step_amplification.png|700px]] | ||
| + | |- | ||
| + | | Dioxygenase (C230) produced upon activation acts on catechol (cat.) to produce yellow output - muconic acid. Catechol is not shown to be produced by cell as it is added by person at arbitrary time. | ||
| + | |- | ||
| + | |[[Image:2-step_amplification.png|700px]] | ||
| + | |- | ||
| + | | The species that are shown in front of vertical line indicating beginning of experiment mean that they have been accumulated beforehand in the cell. TEV protease activates inactive dioxygenase which acts on catechol to produce colour. | ||
| + | |- | ||
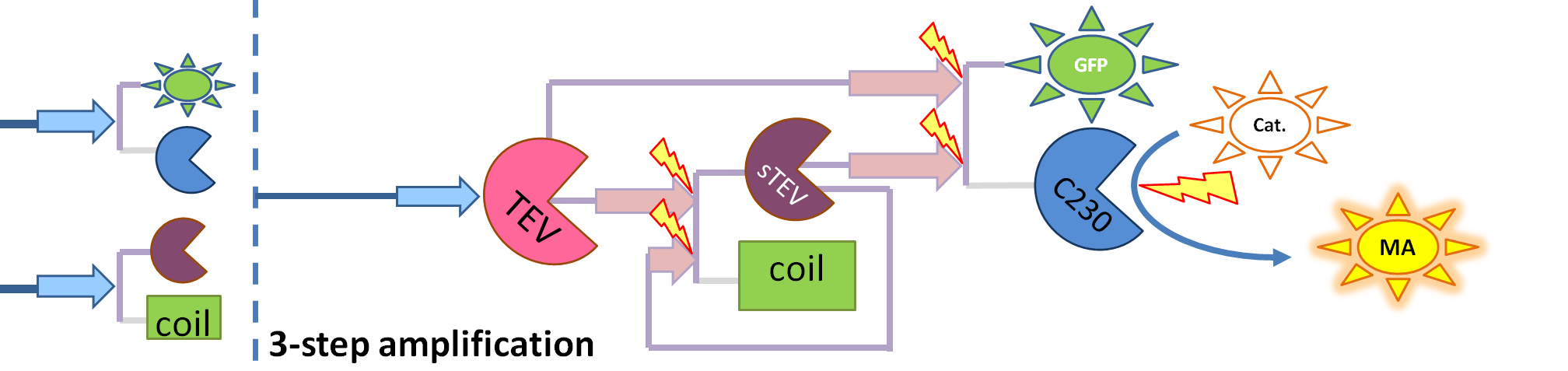
| + | |[[Image:3-step_amplification.png|700px]] | ||
| + | |- | ||
| + | | This diagram introduces inactive split TEV protease attached to coiled-coil as the third amplification step. Both inactive compounds have active site for TEV to act which results in multiple possibilities of action. | ||
|} | |} | ||
</div> | </div> | ||
Revision as of 23:33, 20 October 2010
| Temporary sub-menu: Output Amplification Model; Surface Protein Model; Wet-Dry Lab Interaction; Dry Lab Diary; |
| Introduction to modelling |
In the process of designing our construct two major questions arose which could be answered by computer modelling:
|
| Results & Conclusions | ||
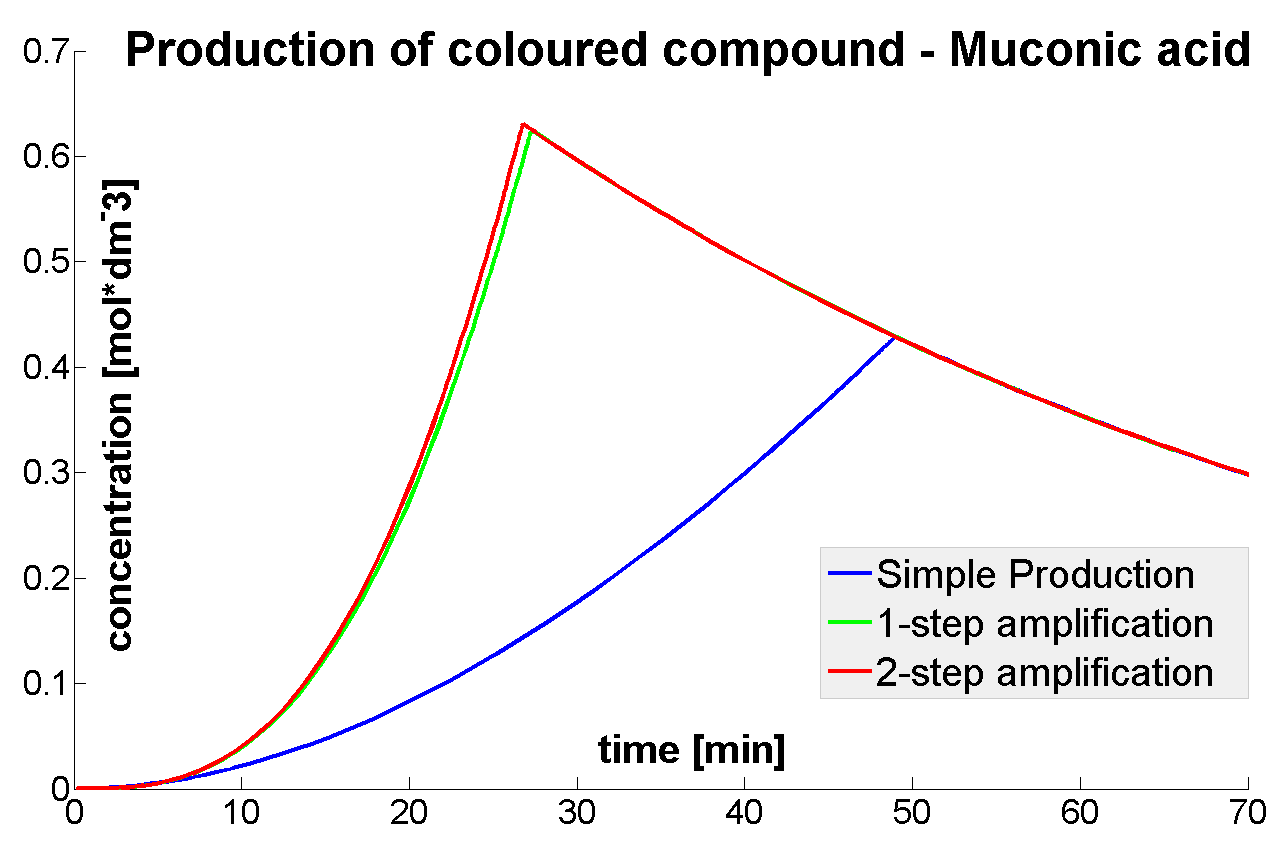
Output Amplification Model
|
 "
"