Team:Freiburg Bioware/NoteBook/Labjournal/October2
From 2010.igem.org
(→qPCRs of virus stocks) |
(→qPCRs of virus stocks) |
||
| Line 2,580: | Line 2,580: | ||
</body> | </body> | ||
| + | </html> | ||
| + | |||
| + | <html xmlns:v="urn:schemas-microsoft-com:vml" | ||
| + | xmlns:o="urn:schemas-microsoft-com:office:office" | ||
| + | xmlns:x="urn:schemas-microsoft-com:office:excel" | ||
| + | xmlns="http://www.w3.org/TR/REC-html40"> | ||
| + | |||
| + | <head> | ||
| + | <meta name="Excel Workbook Frameset"> | ||
| + | <meta http-equiv=Content-Type content="text/html; charset=windows-1252"> | ||
| + | <meta name=ProgId content=Excel.Sheet> | ||
| + | <meta name=Generator content="Microsoft Excel 14"> | ||
| + | <link rel=File-List | ||
| + | href="2010-10-13%20qPCR%20auswertung_html-Dateien/filelist.xml"> | ||
| + | <![if !supportTabStrip]> | ||
| + | <link id="shLink" href="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet001.htm"> | ||
| + | <link id="shLink" href="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet002.htm"> | ||
| + | <link id="shLink" href="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet003.htm"> | ||
| + | |||
| + | <link id="shLink"> | ||
| + | |||
| + | <script language="JavaScript"> | ||
| + | <!-- | ||
| + | var c_lTabs=3; | ||
| + | |||
| + | var c_rgszSh=new Array(c_lTabs); | ||
| + | c_rgszSh[0] = "Tabelle1"; | ||
| + | c_rgszSh[1] = "Tabelle2"; | ||
| + | c_rgszSh[2] = "Tabelle3"; | ||
| + | |||
| + | |||
| + | |||
| + | var c_rgszClr=new Array(8); | ||
| + | c_rgszClr[0]="window"; | ||
| + | c_rgszClr[1]="buttonface"; | ||
| + | c_rgszClr[2]="windowframe"; | ||
| + | c_rgszClr[3]="windowtext"; | ||
| + | c_rgszClr[4]="threedlightshadow"; | ||
| + | c_rgszClr[5]="threedhighlight"; | ||
| + | c_rgszClr[6]="threeddarkshadow"; | ||
| + | c_rgszClr[7]="threedshadow"; | ||
| + | |||
| + | var g_iShCur; | ||
| + | var g_rglTabX=new Array(c_lTabs); | ||
| + | |||
| + | function fnGetIEVer() | ||
| + | { | ||
| + | var ua=window.navigator.userAgent | ||
| + | var msie=ua.indexOf("MSIE") | ||
| + | if (msie>0 && window.navigator.platform=="Win32") | ||
| + | return parseInt(ua.substring(msie+5,ua.indexOf(".", msie))); | ||
| + | else | ||
| + | return 0; | ||
| + | } | ||
| + | |||
| + | function fnBuildFrameset() | ||
| + | { | ||
| + | var szHTML="<frameset rows=\"*,18\" border=0 width=0 frameborder=no framespacing=0>"+ | ||
| + | "<frame src=\""+document.all.item("shLink")[0].href+"\" name=\"frSheet\" noresize>"+ | ||
| + | "<frameset cols=\"54,*\" border=0 width=0 frameborder=no framespacing=0>"+ | ||
| + | "<frame src=\"\" name=\"frScroll\" marginwidth=0 marginheight=0 scrolling=no>"+ | ||
| + | "<frame src=\"\" name=\"frTabs\" marginwidth=0 marginheight=0 scrolling=no>"+ | ||
| + | "</frameset></frameset><plaintext>"; | ||
| + | |||
| + | with (document) { | ||
| + | open("text/html","replace"); | ||
| + | write(szHTML); | ||
| + | close(); | ||
| + | } | ||
| + | |||
| + | fnBuildTabStrip(); | ||
| + | } | ||
| + | |||
| + | function fnBuildTabStrip() | ||
| + | { | ||
| + | var szHTML= | ||
| + | "<html><head><style>.clScroll {font:8pt Courier New;color:"+c_rgszClr[6]+";cursor:default;line-height:10pt;}"+ | ||
| + | ".clScroll2 {font:10pt Arial;color:"+c_rgszClr[6]+";cursor:default;line-height:11pt;}</style></head>"+ | ||
| + | "<body onclick=\"event.returnValue=false;\" ondragstart=\"event.returnValue=false;\" onselectstart=\"event.returnValue=false;\" bgcolor="+c_rgszClr[4]+" topmargin=0 leftmargin=0><table cellpadding=0 cellspacing=0 width=100%>"+ | ||
| + | "<tr><td colspan=6 height=1 bgcolor="+c_rgszClr[2]+"></td></tr>"+ | ||
| + | "<tr><td style=\"font:1pt\"> <td>"+ | ||
| + | "<td valign=top id=tdScroll class=\"clScroll\" onclick=\"parent.fnFastScrollTabs(0);\" onmouseover=\"parent.fnMouseOverScroll(0);\" onmouseout=\"parent.fnMouseOutScroll(0);\"><a>«</a></td>"+ | ||
| + | "<td valign=top id=tdScroll class=\"clScroll2\" onclick=\"parent.fnScrollTabs(0);\" ondblclick=\"parent.fnScrollTabs(0);\" onmouseover=\"parent.fnMouseOverScroll(1);\" onmouseout=\"parent.fnMouseOutScroll(1);\"><a><</a></td>"+ | ||
| + | "<td valign=top id=tdScroll class=\"clScroll2\" onclick=\"parent.fnScrollTabs(1);\" ondblclick=\"parent.fnScrollTabs(1);\" onmouseover=\"parent.fnMouseOverScroll(2);\" onmouseout=\"parent.fnMouseOutScroll(2);\"><a>></a></td>"+ | ||
| + | "<td valign=top id=tdScroll class=\"clScroll\" onclick=\"parent.fnFastScrollTabs(1);\" onmouseover=\"parent.fnMouseOverScroll(3);\" onmouseout=\"parent.fnMouseOutScroll(3);\"><a>»</a></td>"+ | ||
| + | "<td style=\"font:1pt\"> <td></tr></table></body></html>"; | ||
| + | |||
| + | with (frames['frScroll'].document) { | ||
| + | open("text/html","replace"); | ||
| + | write(szHTML); | ||
| + | close(); | ||
| + | } | ||
| + | |||
| + | szHTML = | ||
| + | "<html><head>"+ | ||
| + | "<style>A:link,A:visited,A:active {text-decoration:none;"+"color:"+c_rgszClr[3]+";}"+ | ||
| + | ".clTab {cursor:hand;background:"+c_rgszClr[1]+";font:9pt Arial;padding-left:3px;padding-right:3px;text-align:center;}"+ | ||
| + | ".clBorder {background:"+c_rgszClr[2]+";font:1pt;}"+ | ||
| + | "</style></head><body onload=\"parent.fnInit();\" onselectstart=\"event.returnValue=false;\" ondragstart=\"event.returnValue=false;\" bgcolor="+c_rgszClr[4]+ | ||
| + | " topmargin=0 leftmargin=0><table id=tbTabs cellpadding=0 cellspacing=0>"; | ||
| + | |||
| + | var iCellCount=(c_lTabs+1)*2; | ||
| + | |||
| + | var i; | ||
| + | for (i=0;i<iCellCount;i+=2) | ||
| + | szHTML+="<col width=1><col>"; | ||
| + | |||
| + | var iRow; | ||
| + | for (iRow=0;iRow<6;iRow++) { | ||
| + | |||
| + | szHTML+="<tr>"; | ||
| + | |||
| + | if (iRow==5) | ||
| + | szHTML+="<td colspan="+iCellCount+"></td>"; | ||
| + | else { | ||
| + | if (iRow==0) { | ||
| + | for(i=0;i<iCellCount;i++) | ||
| + | szHTML+="<td height=1 class=\"clBorder\"></td>"; | ||
| + | } else if (iRow==1) { | ||
| + | for(i=0;i<c_lTabs;i++) { | ||
| + | szHTML+="<td height=1 nowrap class=\"clBorder\"> </td>"; | ||
| + | szHTML+= | ||
| + | "<td id=tdTab height=1 nowrap class=\"clTab\" onmouseover=\"parent.fnMouseOverTab("+i+");\" onmouseout=\"parent.fnMouseOutTab("+i+");\">"+ | ||
| + | "<a href=\""+document.all.item("shLink")[i].href+"\" target=\"frSheet\" id=aTab> "+c_rgszSh[i]+" </a></td>"; | ||
| + | } | ||
| + | szHTML+="<td id=tdTab height=1 nowrap class=\"clBorder\"><a id=aTab> </a></td><td width=100%></td>"; | ||
| + | } else if (iRow==2) { | ||
| + | for (i=0;i<c_lTabs;i++) | ||
| + | szHTML+="<td height=1></td><td height=1 class=\"clBorder\"></td>"; | ||
| + | szHTML+="<td height=1></td><td height=1></td>"; | ||
| + | } else if (iRow==3) { | ||
| + | for (i=0;i<iCellCount;i++) | ||
| + | szHTML+="<td height=1></td>"; | ||
| + | } else if (iRow==4) { | ||
| + | for (i=0;i<c_lTabs;i++) | ||
| + | szHTML+="<td height=1 width=1></td><td height=1></td>"; | ||
| + | szHTML+="<td height=1 width=1></td><td></td>"; | ||
| + | } | ||
| + | } | ||
| + | szHTML+="</tr>"; | ||
| + | } | ||
| + | |||
| + | szHTML+="</table></body></html>"; | ||
| + | with (frames['frTabs'].document) { | ||
| + | open("text/html","replace"); | ||
| + | charset=document.charset; | ||
| + | write(szHTML); | ||
| + | close(); | ||
| + | } | ||
| + | } | ||
| + | |||
| + | function fnInit() | ||
| + | { | ||
| + | g_rglTabX[0]=0; | ||
| + | var i; | ||
| + | for (i=1;i<=c_lTabs;i++) | ||
| + | with (frames['frTabs'].document.all.tbTabs.rows[1].cells[fnTabToCol(i-1)]) | ||
| + | g_rglTabX[i]=offsetLeft+offsetWidth-6; | ||
| + | } | ||
| + | |||
| + | function fnTabToCol(iTab) | ||
| + | { | ||
| + | return 2*iTab+1; | ||
| + | } | ||
| + | |||
| + | function fnNextTab(fDir) | ||
| + | { | ||
| + | var iNextTab=-1; | ||
| + | var i; | ||
| + | |||
| + | with (frames['frTabs'].document.body) { | ||
| + | if (fDir==0) { | ||
| + | if (scrollLeft>0) { | ||
| + | for (i=0;i<c_lTabs&&g_rglTabX[i]<scrollLeft;i++); | ||
| + | if (i<c_lTabs) | ||
| + | iNextTab=i-1; | ||
| + | } | ||
| + | } else { | ||
| + | if (g_rglTabX[c_lTabs]+6>offsetWidth+scrollLeft) { | ||
| + | for (i=0;i<c_lTabs&&g_rglTabX[i]<=scrollLeft;i++); | ||
| + | if (i<c_lTabs) | ||
| + | iNextTab=i; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | return iNextTab; | ||
| + | } | ||
| + | |||
| + | function fnScrollTabs(fDir) | ||
| + | { | ||
| + | var iNextTab=fnNextTab(fDir); | ||
| + | |||
| + | if (iNextTab>=0) { | ||
| + | frames['frTabs'].scroll(g_rglTabX[iNextTab],0); | ||
| + | return true; | ||
| + | } else | ||
| + | return false; | ||
| + | } | ||
| + | |||
| + | function fnFastScrollTabs(fDir) | ||
| + | { | ||
| + | if (c_lTabs>16) | ||
| + | frames['frTabs'].scroll(g_rglTabX[fDir?c_lTabs-1:0],0); | ||
| + | else | ||
| + | if (fnScrollTabs(fDir)>0) window.setTimeout("fnFastScrollTabs("+fDir+");",5); | ||
| + | } | ||
| + | |||
| + | function fnSetTabProps(iTab,fActive) | ||
| + | { | ||
| + | var iCol=fnTabToCol(iTab); | ||
| + | var i; | ||
| + | |||
| + | if (iTab>=0) { | ||
| + | with (frames['frTabs'].document.all) { | ||
| + | with (tbTabs) { | ||
| + | for (i=0;i<=4;i++) { | ||
| + | with (rows[i]) { | ||
| + | if (i==0) | ||
| + | cells[iCol].style.background=c_rgszClr[fActive?0:2]; | ||
| + | else if (i>0 && i<4) { | ||
| + | if (fActive) { | ||
| + | cells[iCol-1].style.background=c_rgszClr[2]; | ||
| + | cells[iCol].style.background=c_rgszClr[0]; | ||
| + | cells[iCol+1].style.background=c_rgszClr[2]; | ||
| + | } else { | ||
| + | if (i==1) { | ||
| + | cells[iCol-1].style.background=c_rgszClr[2]; | ||
| + | cells[iCol].style.background=c_rgszClr[1]; | ||
| + | cells[iCol+1].style.background=c_rgszClr[2]; | ||
| + | } else { | ||
| + | cells[iCol-1].style.background=c_rgszClr[4]; | ||
| + | cells[iCol].style.background=c_rgszClr[(i==2)?2:4]; | ||
| + | cells[iCol+1].style.background=c_rgszClr[4]; | ||
| + | } | ||
| + | } | ||
| + | } else | ||
| + | cells[iCol].style.background=c_rgszClr[fActive?2:4]; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | with (aTab[iTab].style) { | ||
| + | cursor=(fActive?"default":"hand"); | ||
| + | color=c_rgszClr[3]; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | function fnMouseOverScroll(iCtl) | ||
| + | { | ||
| + | frames['frScroll'].document.all.tdScroll[iCtl].style.color=c_rgszClr[7]; | ||
| + | } | ||
| + | |||
| + | function fnMouseOutScroll(iCtl) | ||
| + | { | ||
| + | frames['frScroll'].document.all.tdScroll[iCtl].style.color=c_rgszClr[6]; | ||
| + | } | ||
| + | |||
| + | function fnMouseOverTab(iTab) | ||
| + | { | ||
| + | if (iTab!=g_iShCur) { | ||
| + | var iCol=fnTabToCol(iTab); | ||
| + | with (frames['frTabs'].document.all) { | ||
| + | tdTab[iTab].style.background=c_rgszClr[5]; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | function fnMouseOutTab(iTab) | ||
| + | { | ||
| + | if (iTab>=0) { | ||
| + | var elFrom=frames['frTabs'].event.srcElement; | ||
| + | var elTo=frames['frTabs'].event.toElement; | ||
| + | |||
| + | if ((!elTo) || | ||
| + | (elFrom.tagName==elTo.tagName) || | ||
| + | (elTo.tagName=="A" && elTo.parentElement!=elFrom) || | ||
| + | (elFrom.tagName=="A" && elFrom.parentElement!=elTo)) { | ||
| + | |||
| + | if (iTab!=g_iShCur) { | ||
| + | with (frames['frTabs'].document.all) { | ||
| + | tdTab[iTab].style.background=c_rgszClr[1]; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | function fnSetActiveSheet(iSh) | ||
| + | { | ||
| + | if (iSh!=g_iShCur) { | ||
| + | fnSetTabProps(g_iShCur,false); | ||
| + | fnSetTabProps(iSh,true); | ||
| + | g_iShCur=iSh; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | window.g_iIEVer=fnGetIEVer(); | ||
| + | if (window.g_iIEVer>=4) | ||
| + | fnBuildFrameset(); | ||
| + | //--> | ||
| + | </script> | ||
| + | <![endif]><!--[if gte mso 9]><xml> | ||
| + | <x:ExcelWorkbook> | ||
| + | <x:ExcelWorksheets> | ||
| + | <x:ExcelWorksheet> | ||
| + | <x:Name>Tabelle1</x:Name> | ||
| + | <x:WorksheetSource | ||
| + | HRef="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet001.htm"/> | ||
| + | </x:ExcelWorksheet> | ||
| + | <x:ExcelWorksheet> | ||
| + | <x:Name>Tabelle2</x:Name> | ||
| + | <x:WorksheetSource | ||
| + | HRef="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet002.htm"/> | ||
| + | </x:ExcelWorksheet> | ||
| + | <x:ExcelWorksheet> | ||
| + | <x:Name>Tabelle3</x:Name> | ||
| + | <x:WorksheetSource | ||
| + | HRef="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet003.htm"/> | ||
| + | </x:ExcelWorksheet> | ||
| + | </x:ExcelWorksheets> | ||
| + | <x:Stylesheet | ||
| + | HRef="2010-10-13%20qPCR%20auswertung_html-Dateien/stylesheet.css"/> | ||
| + | <x:WindowHeight>12840</x:WindowHeight> | ||
| + | <x:WindowWidth>19440</x:WindowWidth> | ||
| + | <x:WindowTopX>120</x:WindowTopX> | ||
| + | <x:WindowTopY>60</x:WindowTopY> | ||
| + | <x:ProtectStructure>False</x:ProtectStructure> | ||
| + | <x:ProtectWindows>False</x:ProtectWindows> | ||
| + | </x:ExcelWorkbook> | ||
| + | </xml><![endif]--> | ||
| + | </head> | ||
| + | |||
| + | <frameset rows="*,39" border=0 width=0 frameborder=no framespacing=0> | ||
| + | <frame src="2010-10-13%20qPCR%20auswertung_html-Dateien/sheet001.htm" name="frSheet"> | ||
| + | <frame src="2010-10-13%20qPCR%20auswertung_html-Dateien/tabstrip.htm" name="frTabs" marginwidth=0 marginheight=0> | ||
| + | <noframes> | ||
| + | <body> | ||
| + | <p>Diese Seite verwendet Frames. Frames werden von Ihrem Browser aber nicht unterstützt.</p> | ||
| + | </body> | ||
| + | </noframes> | ||
| + | </frameset> | ||
</html> | </html> | ||
Revision as of 09:14, 14 October 2010
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 145 )
- October part 2 (labday 146 - 155 )
- October part 3 (labday 156 - 166 )
- November (labday 167 - 170 )
- Cellculture
146. labday 11.10.2010
Colony PCR of Cloning VP2 Fusion and Super constructs into pSB1C3
Investigator: Achim, Hanna
Comment: Because yesterday's cloning didn't deliver a good separartion of the expected gel bands. Nevertheless ligation and trafo was performed. In order to immediately find out, whether we received successful results a colony PCR will be performed.
Two clones were picked from each plat. In addition to that a positive (pAAV_RC) and a negative control (pSB1C3_lITR) was prepared.
Used primer: 4200 rev and Cap3500 for. Expected fragment size: 885 bp.
PCR was performed following the standard protocol.
All samples match the positive control!
To do: Mini-Prep and sequencing.
Preparation of SDS-PAGE gel (10%)
Investigator: Hanna
Comment: In order to perform a Western Blot of different virus capsids (with and without capsid-motifs), 2 10% SDS polyacrylamid gels were prepared.
Resolving gel: 15 mL
- H2O: 5.9 mL
- Acryl-bisacrylamide mix (30%): 5 mL
- Tris (1.5 M, pH 8.8): 3.8 mL
- SDS (10%): 0.15 mL
- Ammonium persulfate (10%): 0.15 mL
- TEMED: 0.006 mL
Stacking gel (5%): 5 mL
- H2O: 3.4 mL
- Acryl-bisacrylamide mix (30%): 0.83 mL
- Tris (1.5 M, pH 6.8): 0.63 mL
- SDS (10%): 0.05 mL
- Ammonium persulfate (10%): 0.05 mL
- TEMED: 0.005 mL
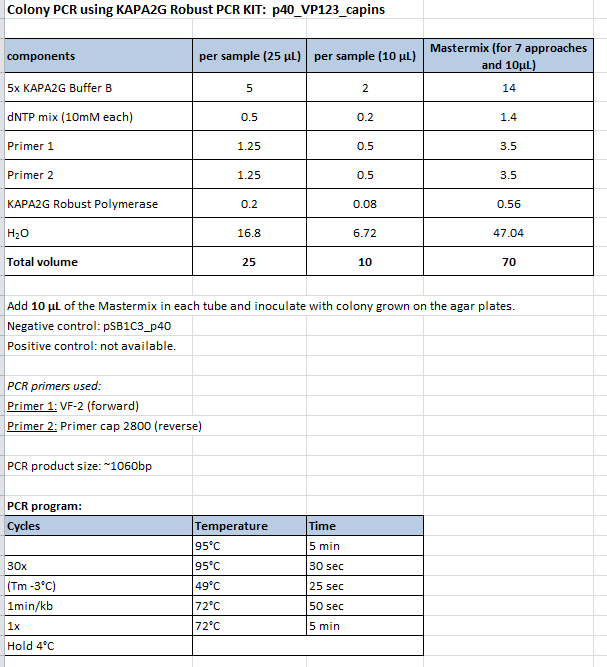
Colony PCR of p40_VP123_capins
Investigator: Bea
Comment: Since the first attempt did not work,and no cells grew on the plate and the same ligation was transformed again into BL-21 and a lot of clones grew on the plate, I decided to perform a colony PCR in order to check several colonies and to inoculate at the same day for a Midi-Prep.
Protocol:
- Primer used: O162
- Primer used: O38
The PCR products were loaded on a 1% agarose gel. The results can be seen above in the gel picture:
Result: We can see two things: The cloning of p40 to VP123 worked quiet well AND the Robust PCR Kit which was used for the first time worked as well.
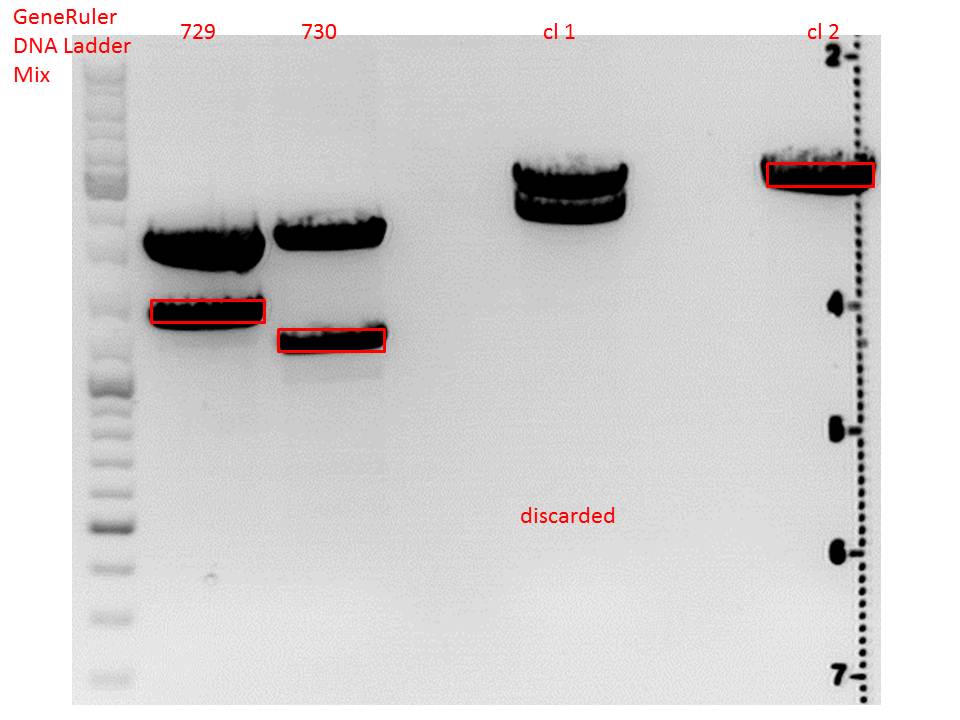
Cloning of lITR_CMV_betaglobin and lITR_phTERT_betaglobin into pSB1C3_CD
Investigator: Stefan
Comment: To produce another GOI for testing in cell culture, the cytosine deaminase needs to be assembled with lITR_promotor_betaglobin. In the next step hgH_rITR needs to be added.
Vector name:
pSB1C3_CD clone 1
pSB1C3_CD clone 2
Insert name:
pSB1C3_lITR_CMV_beta-globin (P729)
pSB1C3_lITR_phTERT_beta-globin (P730)
Digestion:
| components | volume CD clone 1 + 2 /µl | volume P729 /µl | volume P730 /µl |
| DNA | 6 | 14 | 6 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme EcoI | 1 | 1 | 1 |
| Enzyme XbaI | 1 | - | - |
| Enzyme SpeI | - | 1 | 1 |
| H2O | 8 | - | 8 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
CD clone 1 yielded to bands around 2500 bp to 3000 bp. Since the vector was cut only using EcoRI and SpeI, it was expected to be linearized, not to be cut into two fragments this size. Therefore, this sample was discarded and cloning was continued using CD clone 2.
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 729 + CD cl2 | 730 + CD cl2 |
| volume of vector | 3,67 | 2,82 |
| volume of insert | 4,33 | 5,18 |
| T4 ligase buffer (10x) | 1 | 1 |
| T4 ligase | 1 | 1 |
Transformation:
Was performed according to standard protocol using BL21 cells.
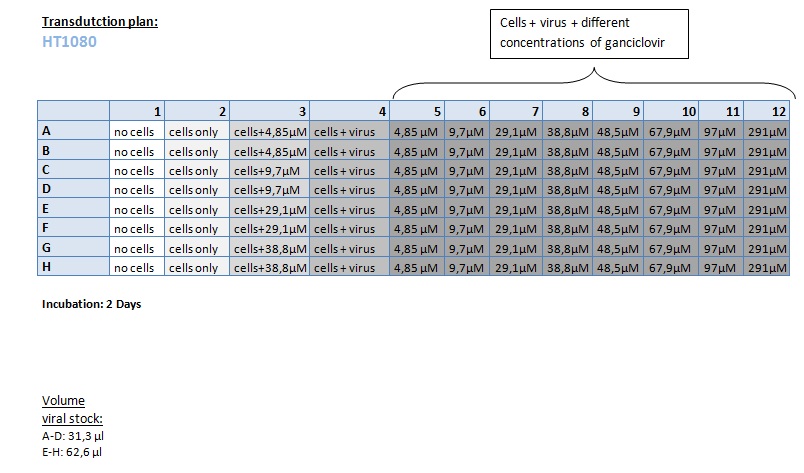
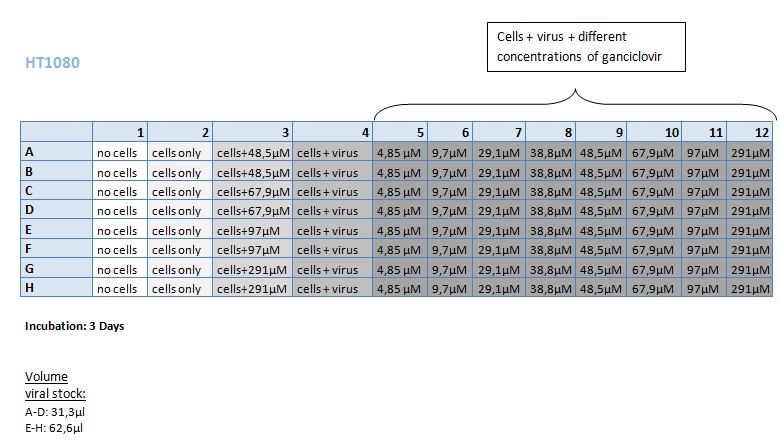
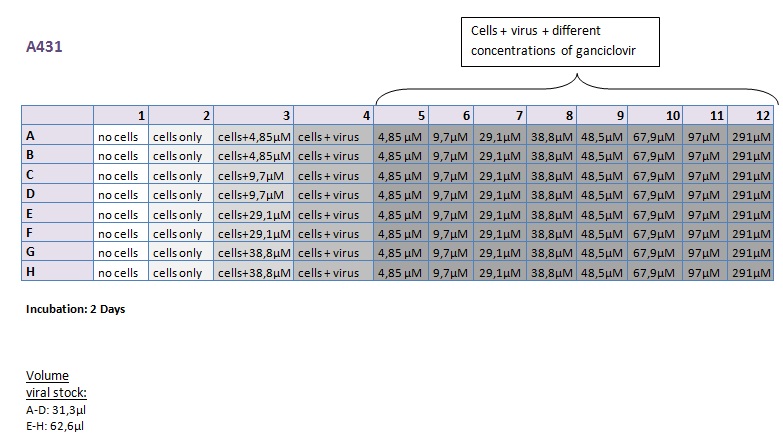
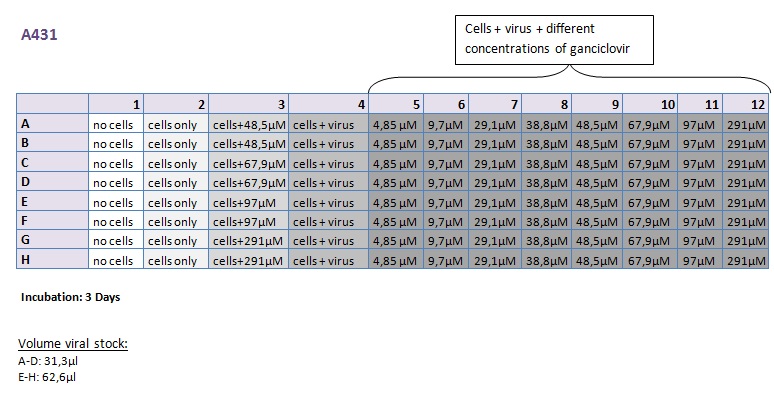
Seeding HT1080 and A431 for testing different concentrations of ganciclovir by MTT-Assay
Investigator: Kerstin, Anissa
- Seeding 4x 96-well plates: 2x HT1080 and 2x A431
FACS-Analysis
Investigator: Kerstin
...
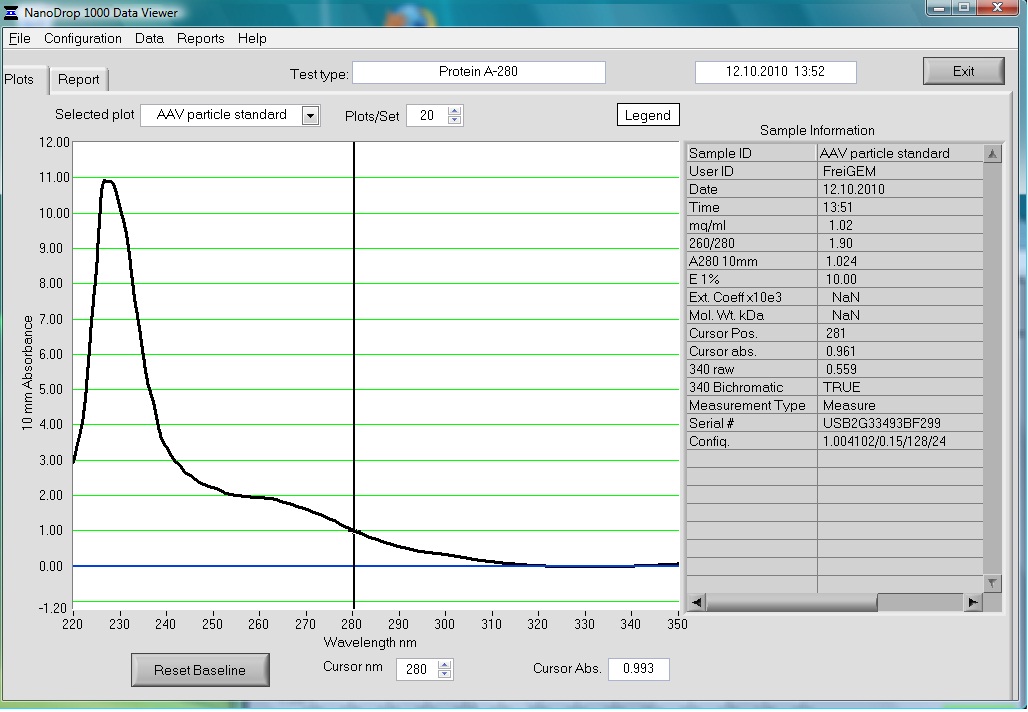
Preparation of the ELISA
Investigator: Volker
The AAV particle standard that contains 3.6x10^9 viral particles was dissolved in 500µl as described in the protocol of the Progen AAV Titration ELISA and a absorption spectrum was measured.
These purified and concentrated viral particles could be used for biophysical measurements, there for the possibility to detect the viral particles by absorption was interesting for us.
The Spectrum measured in the NanoDrop is the following:
Trafo evaluation of Rep52
Investigator:Kira
T4 ligation seems to worked out. The plate contained many colonies.
147. labday 12.10.2010
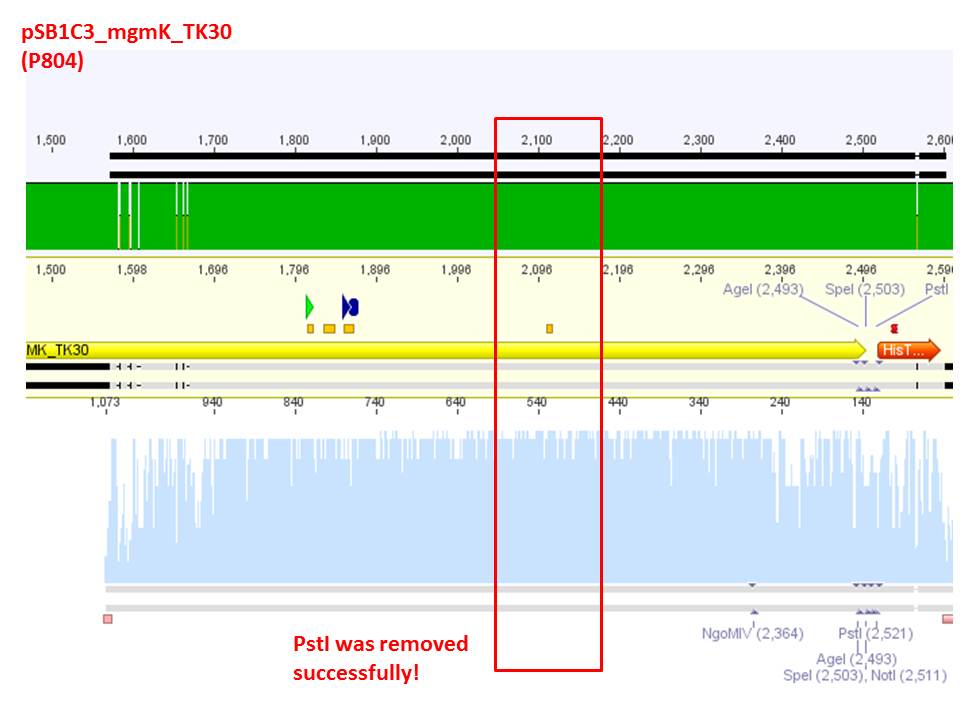
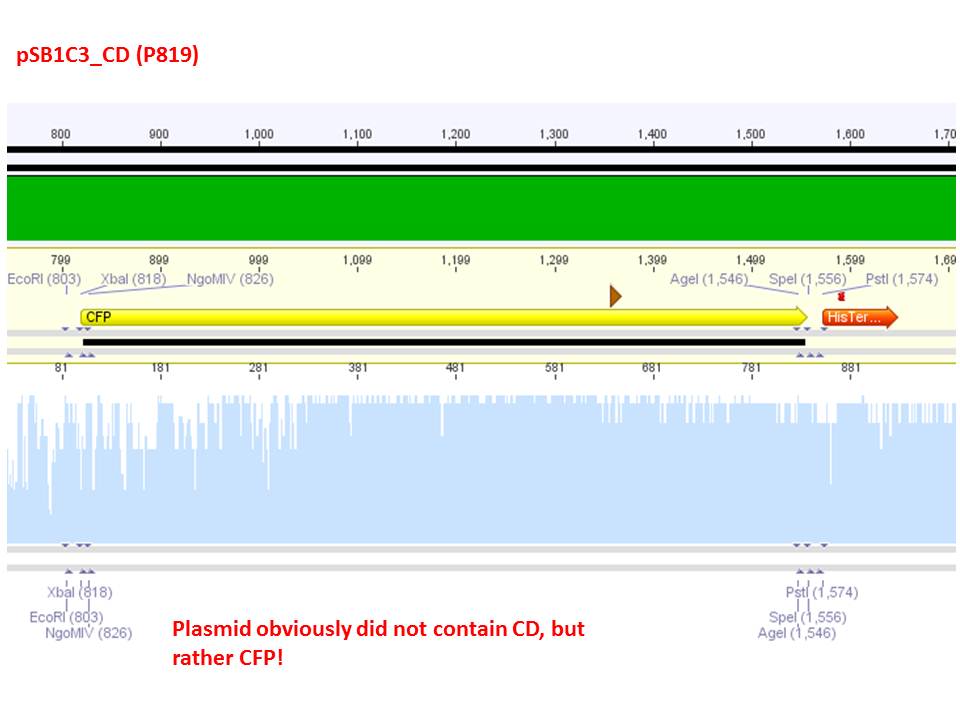
Sequencing analysis: pSB1C3_mGMK_TK30 and pSB1C3_CD
Investigator: Stefan
Test digestion of pSB1C3_CD and pSB1C3_CD_SDM-PstI_new
Investigator: Stefan
Comment: Nachricht von Stefan an Stefan: bitte ergänzen!
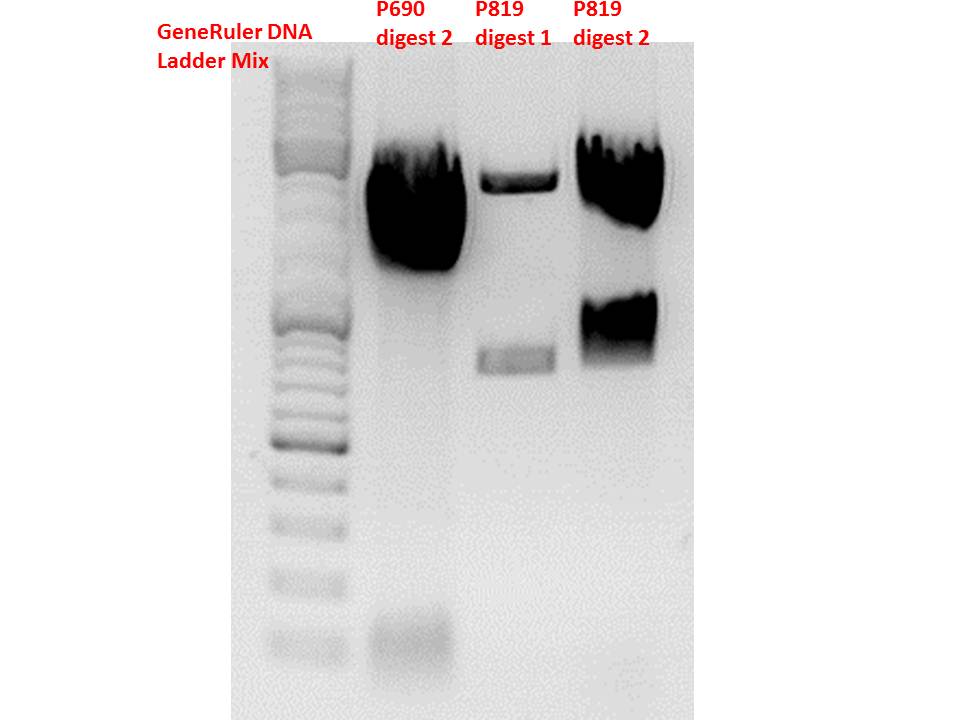
Comment: Sequencing results revealed that there is CFP rather than CD in P819. In order to verify this and exclude the possibility that the wrong plasmid was sent for sequencing test digestions were performed. Additionally the original vector at which the SDM to remove the PstI was performed was digested as well to verify its insert.
| Components | digest 1 / µl | digest 2 / µl |
| DNA | 1 | 7 |
| Buffer 4 | 1 | 1 |
| BSA (10x) | 1 | 1 |
| Enzyme I | SpeI 0,3 | PstI 0,3 |
| Enzyme II | XbaI 0,3 | XbaI 0,3 |
| H2O | 6,4 | 0,4 |
| Total volume | 10 | 10 |
Comment: Using these digestion approaches, it was expected for CFP to deliver always bands at ~750 bp. For the CD it was expected to show bands at 90 bp, 1200 bp and 2000 bp if the CD still contains the PstI restictiton site, 1300 bp and 2000 bp if it was removed.
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: As it can be seen, P819 does contain CFP, therefore a mistakenly sequenced plasmid can be excluded. Also it can be seen that P690 contains the CD. Therefore an additonal SDM of P690 to remove the PstI restriction site is necessary.
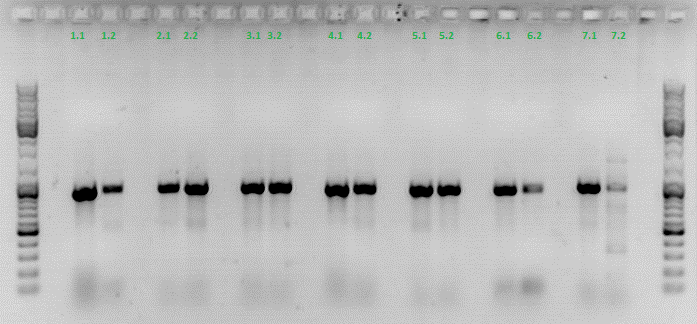
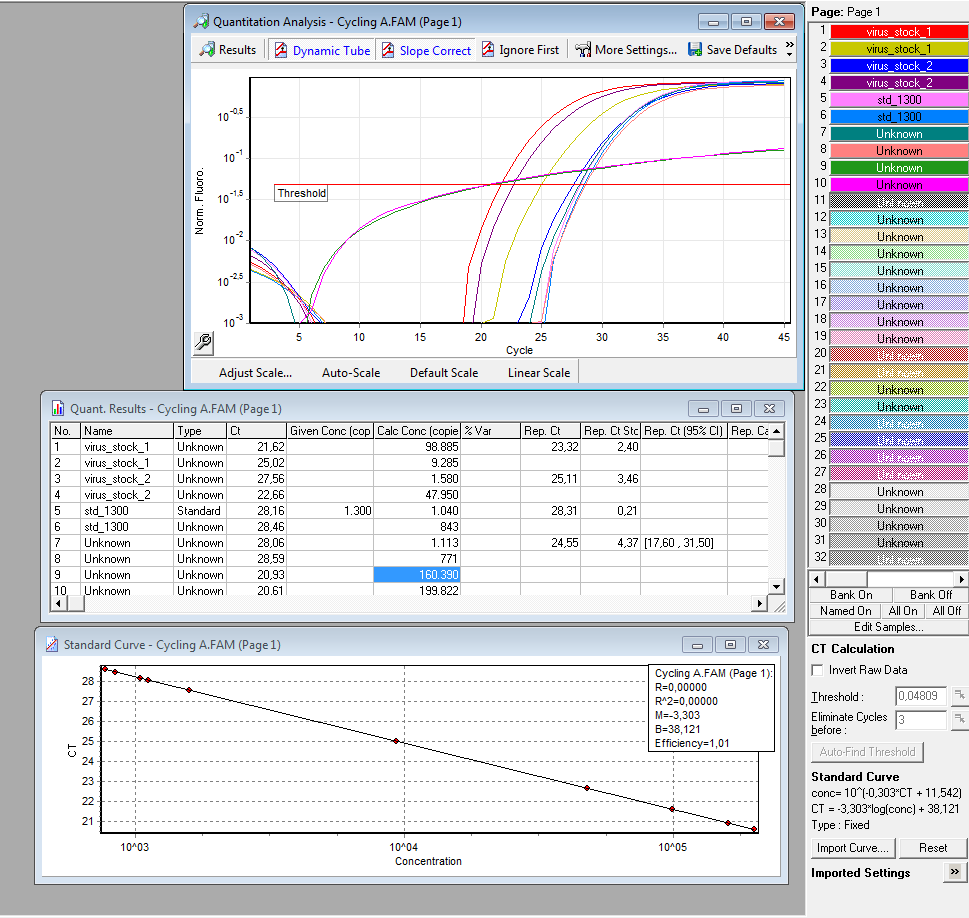
qPCR of virus stock: P816/ TKGMk-Standard; R/C: P468 from 5.10. (VLP harvested by Adrian)
Investigator: Achim
Protocol for quantitative real-time PCR of virus particles
Date: 12.10.
Achim
Virus Stock: A.F.; P816; TKGMK-Standard; R/C: P468; 5.10; consists of supernatant from pelleted cell fragments.
Following the Protocol used by (Rohr et al., 2002).
I digested 5 µl virus dilution with 7,5 µl DNAse I (7,5 units) and 25 µl 50 mM MgCl2 (end concentration 25 mM) in a final volume of 50 µl at 37°C for 30 min.
è DNAse should be heat inactivated at 65°C for 10 min. I forgot that step. The enzyme should however be inactivated in the initial denaturation step.
I prepared the following PCR reactions:
|
Sample |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
9 |
10 |
|
|
1.1 |
1.2 |
2.1 |
2.2 |
S.1 |
S.2 |
NP1 |
NP2 |
MM for samples 1 -8 |
NH1 |
NH2 |
|
PCRmix |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
112.5 |
12.5 |
12.5 |
|
Primer for |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
9 |
- |
- |
|
Primer rev |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
9 |
- |
- |
|
Template |
2 |
2 |
2 |
2 |
5 |
5 |
- |
- |
- |
- |
- |
|
H2O |
8.5 |
8.5 |
8.5 |
8.5 |
5.5 |
5.5 |
10.5 |
10.5 |
- |
12.5 |
12.5 |
|
Total |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
|
25 |
25 |
|
Step |
Time |
Temp |
|
Initial denaturation |
7’ |
95°C |
|
|
|
|
|
Denaturation |
10’’ |
95°C |
|
Annealing/Extension |
30’’ |
60°C |
|
Number of Cycles |
45 |
|
Results:
è Both negative controls containing primers were contaminated with DNA.
è Negative controls containing only H2O were negative.
è Values for plasmids/reaction vary; from 1 – 100 for the same virus stock
è Dilution of the virus solution: 1:10; 2µl PCR sample: 3.2 * 10^5 virus particles/ml
è Because of the contaminated negative controls and the high deviation, I will repeat the experiment tomorrow to verify the results.
References:
Rohr, U., Wulf, M., Stahn, S., Steidl, U., Haas, R., Kronenwett, R., et al. (2002). Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. Journal of virological methods, 106(1), 81-8. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12367732.
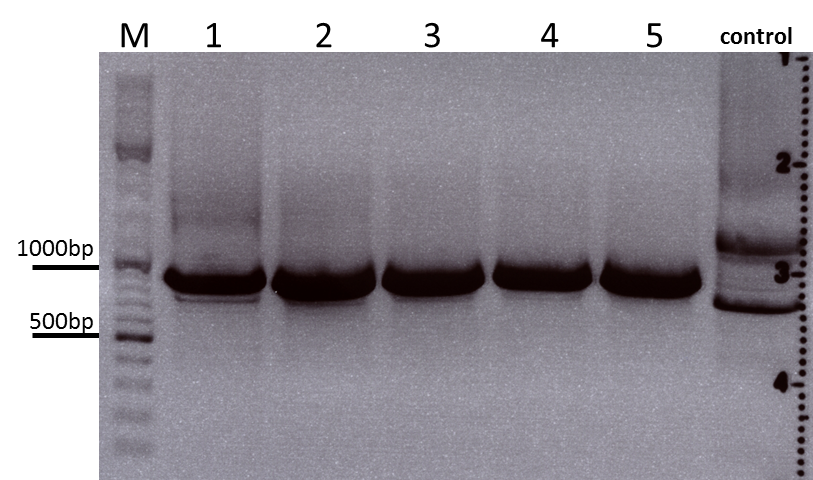
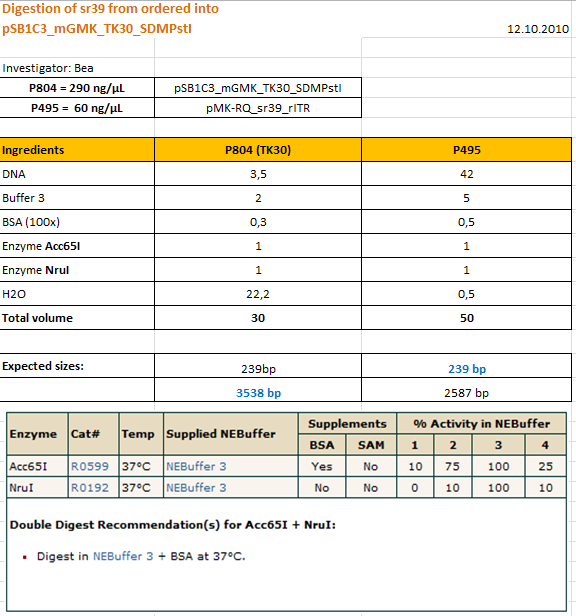
Cloning sr39 into mGMK_TK30
Investigator: Bea
Comment: We do not only want to submit the tk30, a mutant version of the thymidine kinase, but as well another mutant protein which is named sr39. SR39 seems to have a better activity than the tk30 without being fused to the guanylate kinase (mgmk), but the sr39 also works quite well as a fusion protein. We ordered one part of the sr39 and we are now subcloning it into the mgmnk_tk30 construct in order to obtain mgmk_sr39.
Protocol:
The digested fragments were loaded on a 1.2% agarose gel. The results can be seen above in the gel picture: I cut out the small visible band in the right lane, and the upper intensive band of the left lane.
Result:
After gel extraction has been performed, I ligated the two fragments and transformed BL-21 cells, plated them on cm agar plates and incubated them over night at 37°C.
Concentration of virus stock for Western Blot analysis
Investigator: Hanna
For the first Western Blot try, we investigate whether is is enough to concentrate the virus stock (from cell supernatant) and then to simply load it onto a SDS-PAGE gel and perfom immuno-staining.
For this purpose 8 mL virus stock (pHelper, pAAV_RC, GOI=YFP) were concentrated via amicon filtration.
- First, amicon filter was washed with 4 mL PBS.
- Then 4 mL virus stock was loaded onto the filter and centrifuged at 3000 rpm for 15 minutes.
- Flow-through was discarded and additional virus stock was loaded, centrifuged at 3000 rpm for 25 minutes.
- Flow-through was discarded, remaining solution was re-suspended and additional virus stock was added. Amicon filter was centrifuged at 3000 rpm for 50 minutes.
- Flow-through was discarded. Residual protein solution: 500 µL. Filter was washed with 3 mL PBS, centrifuging 35 minutes two times.
- 2 x 40 µL protein solution (16x) was taken and mixed with Laemmli buffer and stored over night @ 4°C.
To do: SDS-PAGE and Western Blot tomorrow.
Perfomance of another try by harvesting viruses of the cell pellet with RIPA buffer & freeze/ thaw.
Western Blot of unmodified, and modified capsids (N-terminal fusion and VP1 insertion with CFP ref. mVenus) --> size shift should be detectable.
Transduction of HT1080 and A431
Investigator: Kerstin
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B651 = pSB1C3_P40_VP123
- B652 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR
- B653 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR
- B654 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B655 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B656 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B657 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B658 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B659 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B660 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR
- B661 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR
Mini-Prep was performed according to standard protocol:
- P821 = pSB1C3_P40_VP123 c = 327,4 ng/µl
- P822 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 342,9 ng/µl
- P823 = pSB1C3_lITR_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 324,0 ng/µl
- P824 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 81,6 ng/µl
- P825 = pSB1C3_lITR_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 148,4 ng/µl
- P826 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 105,8 ng/µl
- P827 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 121,1 ng/µl
- P828 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 188,4 ng/µl
- P829 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 140,4 ng/µl
- P830 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR c = 197,2 ng/µl
- P831 = pSB1C3_lITR_phTERT_ß-globin_CFP_hgH-rITR c = 207,3 ng/µl
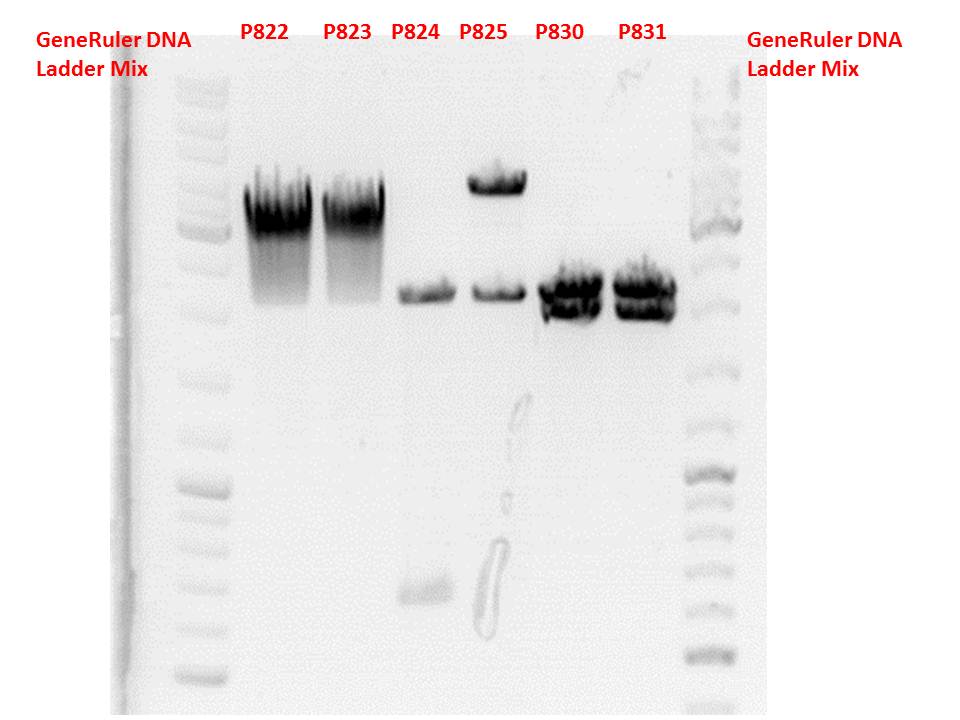
Test digestion:
| Components | P822 + P823 / µl | P824 + P825 / µl | P830 + P831 / µl |
| DNA | 1,5 | 1,5 | 2 |
| Buffer 4 | 1 | 1 | 1 |
| BSA (10x) | 1 | 1 | 1 |
| PstI | 0,3 | - | - |
| XbaI | 0,3 | 0,3 | 0,3 |
| SpeI | - | 0,3 | 0,3 |
| H2O | 5,9 | 5,9 | 5,4 |
| Total volume | 10 | 10 | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: P822 and P823 smear a lot, therefore evaluation is difficult. Another test digestion would be reasonable.
P824 contains hgH_rITR, therefore it will be discarded. P825 looks well and was sent for sequencing.
P830 and P831 look well and will be sent for sequencing tomorrow.
Repetiton: Site-directed mutagenesis in pSB1C3_CD construct
Investigator: Stefan
Comment: Sequencing revealed that there is no CD in our pSB1C3_CD but rather CFP. So, anonther approach of the SDM needs to be performed.
Protocol using the QuickChange Lightning Kit from Stratagene:
Ingredients:
| Ingredients | Volume / µl |
| 10x reaction buffer | 2,5 |
| dNTP | 0,5 |
| forward primer: O191 | 0,56 |
| reverse primer: O192 | 0,57 |
| DNA Template | 0,5 (1:10 dilution) |
| QuikSolution Reagent | 0,75 |
| QuikChangeLightning Enzyme | 0,5 |
| H2O | 14,35 |
| Total volume | 25 |
The following PCR program was used:
- 95 °C 120 s
- 95 °C 20 s
- 60 °C 60 s
- 68 °C 105 s
- Goto step 2 repeat 17 times
- 68 °C 300 s
Digestion using DpnI:
To digest methylated and hemi-methylated DNA, 1 µl of DpnI was added and the mixture was incubated for 10 minutes at 37 °C.
Transformation:
Trafo was performed according to protocol, but using XL1b cells instead of XL-10 Gold cells.
148. labday 13.10.2010
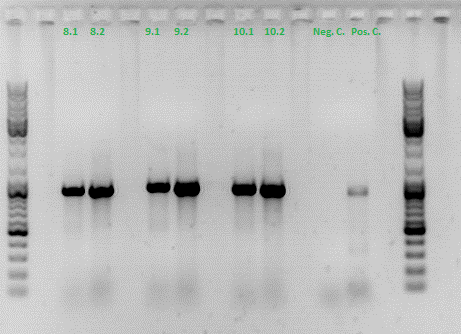
qPCRs of virus stocks
Investigator: Achim
|
Sample: |
Plasmid, Glycerol stock: |
Sample No. |
|
pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10 clone 1 |
B561 |
1 |
|
pSB1C3_001_RC_IRCK_P5tataless clone1 |
P668 |
2 |
|
pSB1C3_lITR_CMV_beta-globin_mVenus_rITR clone1 |
P378 |
3 |
|
pSB1C3_lITR_CMV_mVenus_hGH_rITR clone1 |
P377 |
4 |
|
pAAV_RC_RepCapIns_SDMKpnI |
P468 |
5 |
Digestion: 37°C, 30 min
|
|
1 |
2 |
3 |
4 |
5 |
MM x6 |
|
Virus solution |
5 |
5 |
5 |
5 |
5 |
|
|
DNAseI (7,5 U) |
7,5 |
7,5 |
7,5 |
7,5 |
7,5 |
45 |
|
MgCl2 50 mM |
5 |
5 |
5 |
5 |
5 |
30 |
|
H2O |
32.5 |
32.5 |
32.5 |
32.5 |
32.5 |
195 |
|
Total |
50 |
50 |
50 |
50 |
50 |
300 |
PCR reaction:
|
Sample |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
15 |
16 |
|
|
1.1 |
1.2 |
2.1 |
2.2 |
3.1 |
3.2 |
4.1 |
4.2 |
5.1 |
5.2 |
S.1 |
S.2 |
NP1 |
NP2 |
MM for samples 1 -14 (x15) |
NH1 |
NH2 |
|
PCRmix |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
12.5 |
187,5 |
12.5 |
12.5 |
|
Primer for 1:10 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
15 |
- |
- |
|
Primer rev 1:10 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
15 |
- |
- |
|
Template |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
5 |
5 |
- |
- |
- |
- |
- |
|
H2O |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
8.5 |
5.5 |
5.5 |
10.5 |
10.5 |
- |
12.5 |
12.5 |
|
Total |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
|
25 |
25 |
 "
"