Team:TU Munich/Project
From 2010.igem.org
| Line 33: | Line 33: | ||
| - | + | ODER: | |
| + | |||
| + | |||
= Our vision = | = Our vision = | ||
<div align="justify"> | <div align="justify"> | ||
Revision as of 13:57, 2 October 2010
|
||||||||||||||||
|
|
VisionAlthough classical molecular biology and genetic engineering equipped the science community with many functional proteins and possible applications, usage and linking different parts together to a working network still requires highly complicated strategies. Often the assembly of different biological parts tends to be error-prone and very difficult to extend since only few switches are established and studied in a way that they deliver reliable basic units. Even those like the ultimate protein expression control unit, the lac operon, are limited: With the lac operon only 0 and 1 can be achieved and only once per expression construct and cell. Our approach is to change that and to enable new and more exact ways to control E. coli cells using RNA-RNA-interaction based switches (bioLOGICS).
ConceptThe basic principle of our switches are short RNA sequences, the scientific idea shares similiarities with the principle of antitermination but also inherits a completely new way of RNA based transcription regulation. We used three-dimensional structure predictions and thermodynamicaly calculation to develop a set of switches (about 50 bases) and signals (about 20 bases). The switch forms a stem loop which can be resolved upon binding of a signal. On/Off switching is therefore possible delivering an easy and flexible way to control gene transcription.
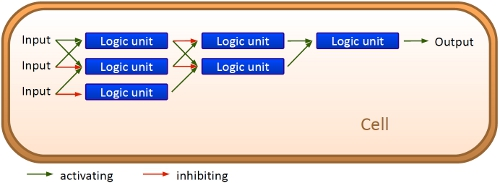
Upscaling: Building networksWith those short RNA sequences as the basic unit, a network can be easily constructed. And and or-circuits can be constructed based on this on/off signal. Since the number of theoretical switch/signal pairs is nearly endless with the basic unit being of a very small size, upscaling is no problem: RNA signal can be produced in response to an RNA signal binding to an RNA switch. This may be one of the major differences to ribozyme and especially protein based networks. While the complexicity of protein-protein interactions may work for cells, constructing networks without just copying complete operons is hardly possible. With the small size of our units 10 switch/signal pairs occupy the space of an average protein on a plasmid. Circuits based on bioLOGICS may play a key role for gene regulation with more variations than just on/off.
Work ProgressEvery network starts with a basic unit. While our declared aim is to enable networks allowing fine-tuning of gene expression beyond the regular on/off the first tests starts with exploring such an on/off switch/signal pair. We tested its efficiency, robustness and reproducibility in vivo, in vitro and in silico. Furthermore we developed a software which allows easy constructions of networks delivering a ready network.
Abstract
By developing a computational biological network based on RNA logical devices we will offer everyone the opportunity to 'program' their own cells with individual AND/OR/NOT connections between BioBricks of their choice. Thereby, BioBricks can finally fulfill their original assignment as biological parts that can be connected in many different ways. We will achieve this by engineering simple and easy-to-handle switches based on predictable RNA/RNA-interactions regulating transcriptional termination. These switches represent a complete set of logical functions and are capable of forming arbitrarily complex networks.
ODER:
Our visionThe idea behind our project is to change the way BioBricks have been used up to now. Over the years, many receptors and signals have been constructed as BioBricks during the annual iGEM competition, but still it is not possible to interconnect these Bricks in a complex biological network resuting in a cell, that is able to respond to its environment giving differenciated responses depending on the input signals. (Beispiel: cambridge hat das gemacht, xx dies, aber eine zelle kann nicht beides... Over the years, many teams participating in the iGEM competition spent their time on constructing receptors and systems to detect a certain input that a variety of gorgeous oppurtunities is available so far. Nevertheless, until now it is not possible to link all those functionalities and build up a network giving differenciated responses to several of those input signals, where the molecular response depends on the complex composition of the environment a cell faces. We would like to offer this possibility to everyone.
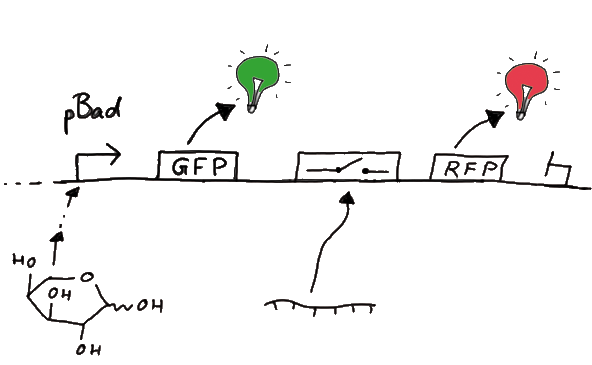
The concept we rely on for our design of RNA-switches is based on the principle of attenuation. ExperimentsWe designed several experiments to test our switches, all of them based on fluorescence measurements. We designed experiment setting for measurements in vivo as well as in vitro. Our in vitro measurements relied on two different experiment set-ups. While the first was based on a commercial E. coli-lysate, the latter was reporting on a transcriptional level only, eliminating most of the possible side-effects one could expect in the complex behaviour of a living cell or cell-lysate. Read more ResultsWe ...blablabla Read more thing to movebioLOGICS: Logical RNA-Devices Enabling BioBrick-Network Formation Abstract Among the goals of iGEM is the creation of synthetic biological parts and their utilization to achieve novel features and behavior in biological systems. The emphasis of our project is put on this latter, "systems" aspect of iGEM. More precisely, we aim at the development and experimental demonstration of a scalable approach for the realization of logical functions in vivo. By developing a computational biological network based on RNA logical devices we will offer everyone the opportunity to 'program' their own cells with individual AND/OR/NOT connections between BioBricks of their choice. Thereby, BioBricks can finally fulfill their original assignment as biological parts that can be connected in many different ways. We will achieve this by engineering simple and easy-to-handle switches based on predictable RNA/RNA-interactions regulating transcriptional termination. These switches represent a complete set of logical functions and are capable of forming arbitrarily complex networks. The ExperimentsFluorescent proteins as reporterOur initial idea to prove our concept of antitermination was to use flourescent proteins as reporters. This approach gives the opportunity to measure the termination and antitermination efficiency of our designed BioBricks in vivo as well as in vitro, the latter using a translation kit based on e.coli lysate.
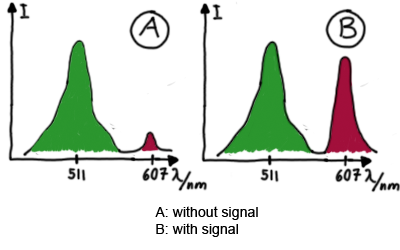
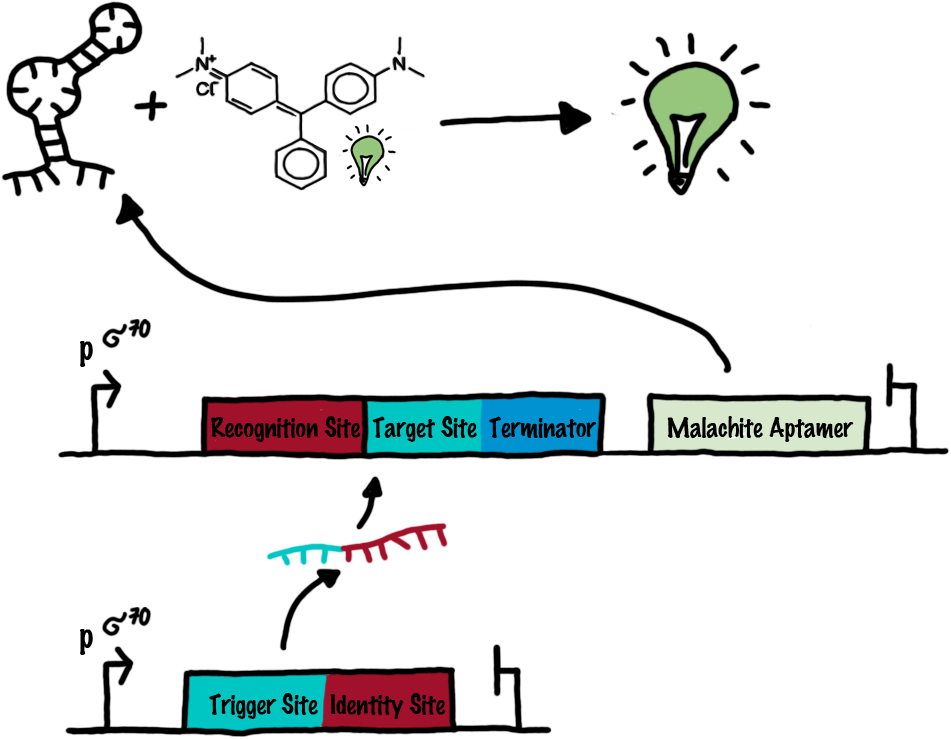
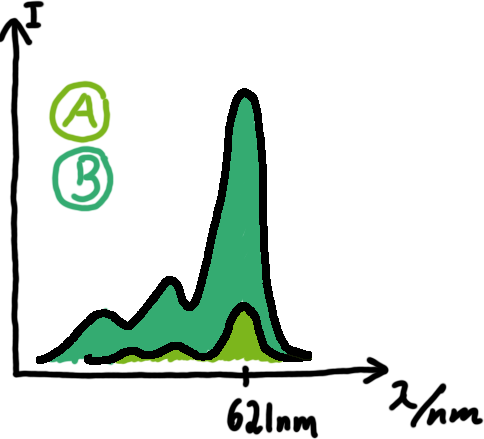
Wiith these measurements, it should also be possible to observe differences in efficiency of termination as well as antitermination between our designed switches. Measurements with the malachite green aptamer as reporterA second possibility to measure parameters of our switches we came up with, was the idea to investigate our system on the transcriptional level only. Therefore, we decided to use malachite green as reporter. Malachite green in a fluorescent dye, whose emission increasing dramaticly (about 3000 times) upon binding of a specific RNA-aptamer. ---concept to be desribed, as well as literature---
<ref>refs</ref>
Also we made constructs, where the transcription of the signal-RNA is under the control of a sigma(70) promoter. These two linear DNA-constructs, together with the e.coli RNA-polymerase and the right buffer conditions should represent an easy-to-handle measurement kit on the transcriptional level.
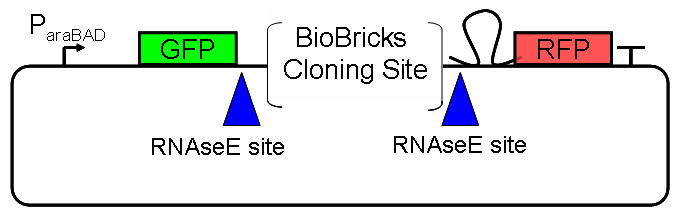
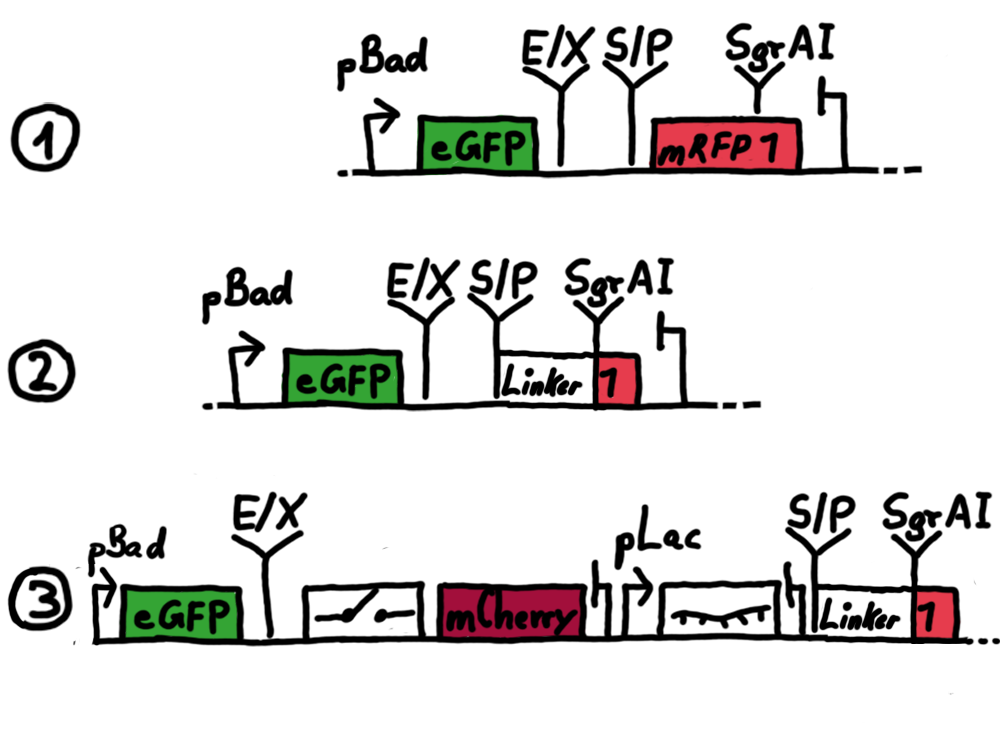
Results Flourescent proteinsUnfortunatly, we had to change the reporter construct two times during our experiments as several problems occured in our measurements: First Try: based on the measurement plasmid pSB1A10At the beginning, we decided to use the reporter plasmid [http://partsregistry.org/Part:pSB1A10 pSB1A10] from the registry. It consists of the fluorescent proteins eGFP and mRFP1. Each sequence includes a ribosome binding site and a stop-codon; the two genes are divided by a cloning side including the BioBrick cleavage sites.In front of the eGFP sequence, the plasmid includes an arabinose-inducable promoter. The plasmid also contains an ampicilline resistence. We cloned our switches into the cloning site of the measurement plasmid and used an empty cloning site as control; our signal-RNAs we cloned into the [http://partsregistry.org/Part:pSB1K3 pSB1K3] vector, together with the BioBricks R0011 (Lac promoter) and B0014 (double terminator of transcription). Afterwards, we cut pSB1K3 with Aat2 and Pst1 and pSB1A10 with Nsi1 and Aat2 and ligated those fragments of each plasmid that contained our Bricks to get a Monsterplasmid. We transformed BL21(DE3) cells with the plasmid. We set up cultures, induced the arabinose promoter and measured the GFP and mRFP1 excitation/emission spectra within time.
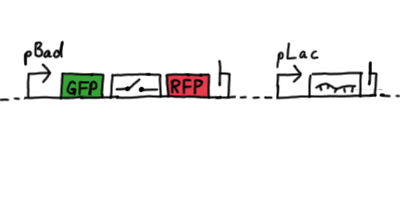
Second Try: A measurement plasmid of our own designTo design our own plasmid to overcome the problems that occurred in our first try gave us tghe possibility to overcome several other problems: Third Try: One promoter for each proteinWe decided to use the measuremnt plasmid we developed in our second try but to clone another L-arabinose induced promoter into the plasmid, in front of our switch followed by the mCherry sequence. In this way, we still can use GFP fluorescence as internal control, because both protein transcription is under the control of a promoter of identical design. Though we are still not able to tell exactly why our previous measurements did not work, but with this construct we measured the first time fluorescence of the mCherry protein in our positive control.
This is a template page. READ THESE INSTRUCTIONS.
You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples HERE.
You MUST have a team description page, a project abstract, a complete project description, a lab notebook, and a safety page. PLEASE keep all of your pages within your teams namespace.
|
|||||||||||||||
 "
"