Team:SDU-Denmark/labnotes9
From 2010.igem.org
(→Flagella Group) |
(→Ligation of FlhDCmut and pSB1AK3) |
||
| Line 413: | Line 413: | ||
'''Results:''' <Br> | '''Results:''' <Br> | ||
<Br><Br> | <Br><Br> | ||
| + | === Colony PCR of FlhDCmut in pSB1AK3 and pSB1C3 === | ||
| + | <br> | ||
| + | '''Date:''' 9/15 2010<Br> | ||
| + | '''Done By:''' Louise<Br> | ||
| + | '''Protocol:''' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3 CP1.3]<Br> | ||
| + | '''Notes:''' We made 8 C-PCRs with colonies transformed with pSB1AK3 and 7 C-PCRs with colonies transformed with pSB1C3 (we made 8 but due to a mishab sample 5 was not run. Taq polymerase, VF2- and VR primers were used. Only Elongation time was altered from 2 min in the protocol to 1.5 min<Br> | ||
| + | '''Results:''' <Br> | ||
| + | The picture first picture is of FlhDCmut in pSB1AK3. This shows one band at about 400bp, the double terminator. This indicates that non of the colonies had the right plasmid.<br> | ||
| + | The second picture is of FlhDCmut in pSB1C3.This shows the 4 of the 7 colonies had plasmids containing the right incert (sample 1,3, 7 and 8). These samples show a band at approximately 1200bp (FlhDCmut in pSB1C3 is 1248bp). the three other samples show a band heavier than 2000bp <br> | ||
| + | [[Image:Team SDU-Denmark FlhDCmut indsat 1. gang i 1AK3 og 1C3.jpg|300px]] | ||
| + | <br> | ||
| + | |||
| + | ---- | ||
== Photosensor group == | == Photosensor group == | ||
Revision as of 10:07, 1 October 2010
Lab notes (9/6 - 9/12)
Flagella Group
Two-step PCR of miniprep
First step: PCR of miniprep with mutation primers
Done by: Louise
Date: September 7th
Protocol: [CP1.1]
Notes:
3 x Premix 1:
114.3ul water
15ul Pfu Buffer
4.5ul dNTP
3ul MgSO4
4.5ul FlhDC fw
4.5ul FlhDCmut rev
1.2ul PFU
3ul template (miniprep)
3 x Premix 2:
114.3ul water
15ul Pfu Buffer
4.5ul dNTP
3ul MgSO4
4.5ul FlhDCmut fw
4.5ul FlhDC rev
1.2ul PFU
3ul template (miniprep)
Results:
NanoDrop:
Sample 1.1: Concentration: 359.5ng/ul Purity: 1.83/2.20
Sample 2.1: Concentration: 304.1ng/ul Purity: 1.85/2.23
The PCR samples were run on a 1.5% gel with a 100bp-1000bp ladder. The first three samples are all positioned between 100bp and 200bp, this is the part of the gene run with FlhDCmut fv and FlhDC rev primers. The other three samples are all positioned betveen 900bp and 1000bp, this is the pcr with FlhDC fv and FlhDCmut rev primers. All samples looked okay and were pooled as sample 1.1 (light band) and sample 2.1 (heavy band)
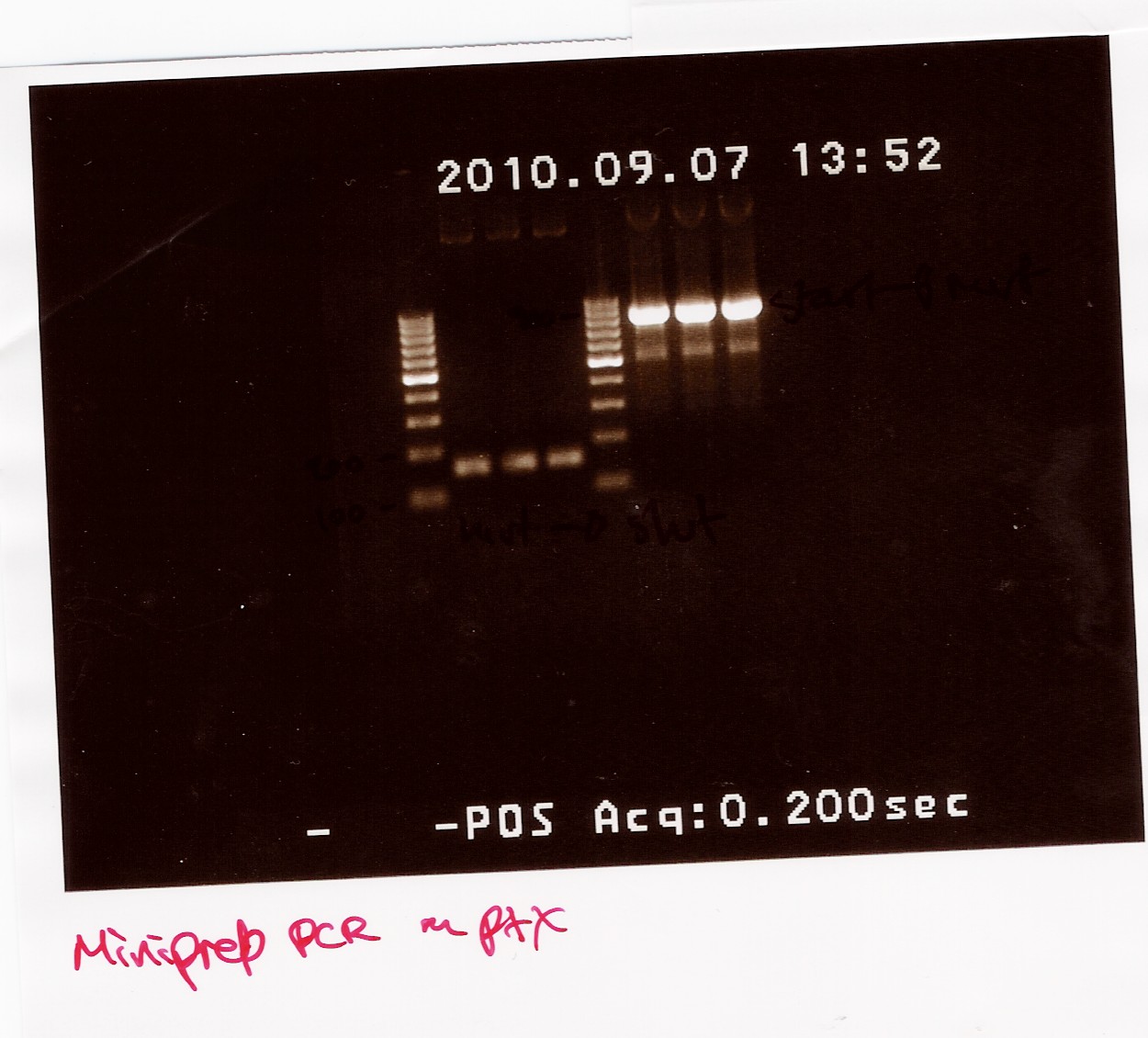
A gel extraction of the PCR product of sample 1.1 and 2.1 were made:

The gel extraction products were used in the preceding PCRs.
Second step: 2-step PCR with mutated template sample 1.1 and 2.1 and FlhDC fw and rev primers
Done by: Maria
Date: September 9th
Protocol: [CP1.1]
Notes:
Premix:
38ul water
5ul PFU buffer + MgSO4
1.5ul dNTP
1ul Sample 1.1
1ul Sample 2.1
1.5ul FlhDC fw primer
1.5ul FlhDC rev primer
0.5ul PFU
PCR Program:
| 1:Start |
95C |
2 min |
| 2: Denaturing |
95C |
30 sec |
| 3: Annealing |
56C |
30 sec |
| 4: Elongation |
72C |
2 min |
| 5: |
GO TO |
2 rep. 4x |
| 6: Denaturing |
95C |
30 sec |
| 7: Annealing |
63C |
30 sec |
| 8: Elongation |
72C |
2 min |
| 9: |
GO TO |
6 rep. 25x |
| 10: End |
72C |
5 min |
| 12: Hold |
4C |
Results:
Some of the PCR product was run on a 1.5% gel with a 10kb ladder.
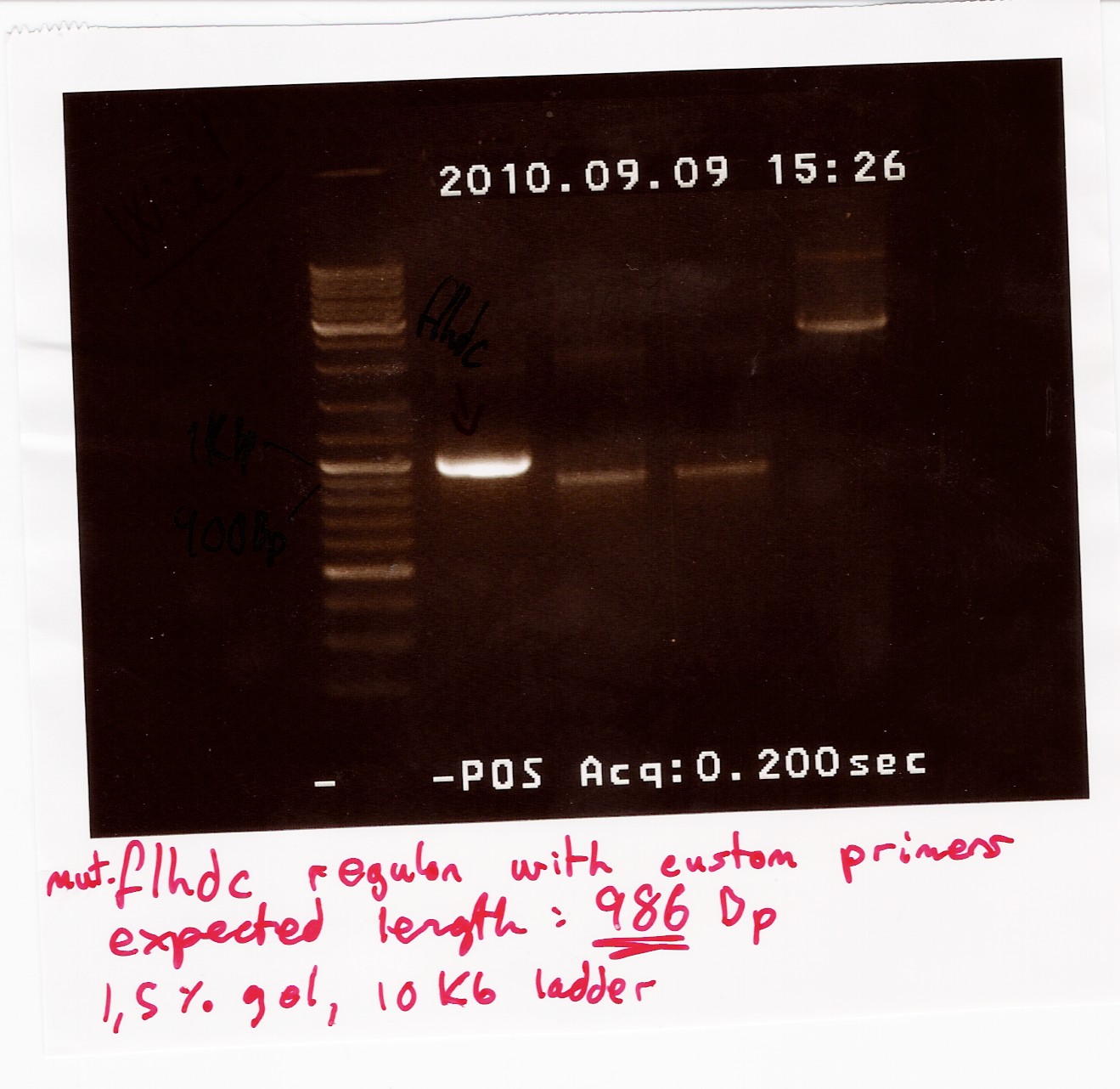
The rest of the product was extracted from a new gel, unfortunately there were problems with the camera, so there is no picture of the gel extraction.
PCR of Gel extraction
Done by: Maria
Date: September 9th
Protocol: [CP1.1]
Notes:
Premix x 6:
234ul water
30ul PFU buffer + MgSO4
9ul dNTP
9ul FlhDC fw primer
9ul FlhDC rev primer
2.5ul PFU
6ul template
| 1:Start |
95C |
2 min |
| 2: Denaturing |
95C |
30sec |
| 3: Annealing |
63C |
30 sec |
| 4: Elongation |
72C |
2 min |
| 5: |
GO TO |
2 rep. 29x |
| 6: End |
72C |
5 min |
| 7: Hold |
4C |
Results:
The PCR worked fine, unfortunately there were problems with the camera, so there is no picture of the gel.
Restriction Digest of Gel Extracted FlhDCmut, pSB1C3 and pSB1AK3
Date: 9/12 2010
Done By: Sheila
Protocol: RD1.1
Notes: The restriction digests are made in preperation for two different ligations, the first is the ligation of FlhDCmut into the submission backbone; pSB1C3. The second is the the ligation of FlhDCmut into pSB1AK3. Different restriction enzymes were used in, as shown in the following tables
Restriction Digest mixtures
FlhDCmut
| H2O | 48μL |
| EcoR1 | 4μL |
| Pst1 | 4μL |
| FD Green buffer | 8μL |
| Sample | 20μL |
pSB1C3
| H2O | 24μL |
| EcoR1 | 2μL |
| Pst1 | 2μL |
| FD Green buffer | 4μL |
| Sample | 10μL |
FlhDCmut and pSB1AK3 Restriction digest
FlhDCmut
| H2O | 48μL |
| EcoR1 | 4μL |
| Spe1 | 4μL |
| FD Green buffer | 8μL |
| Sample | 20μL |
pSB1AK3
| H2O | 24μL |
| EcoR1 | 2μL |
| Xba1 | 2μL |
| FD Green buffer | 4μL |
| Sample | 10μL |
Following digestion, the samples were loaded and run on a 1.5% agarose gel, before being extracted prior to ligation.
Results:
Ligation of FlhDCmut and pSB1C3
Date: 9/12 2010
Done By: Sheila
Protocol: LG1.2
Notes:
Results:
Ligation of FlhDCmut and pSB1AK3
Date: 9/12 2010
Done By: Sheila
Protocol: LG1.2
Notes:
Results:
Colony PCR of FlhDCmut in pSB1AK3 and pSB1C3
Date: 9/15 2010
Done By: Louise
Protocol: CP1.3
Notes: We made 8 C-PCRs with colonies transformed with pSB1AK3 and 7 C-PCRs with colonies transformed with pSB1C3 (we made 8 but due to a mishab sample 5 was not run. Taq polymerase, VF2- and VR primers were used. Only Elongation time was altered from 2 min in the protocol to 1.5 min
Results:
The picture first picture is of FlhDCmut in pSB1AK3. This shows one band at about 400bp, the double terminator. This indicates that non of the colonies had the right plasmid.
The second picture is of FlhDCmut in pSB1C3.This shows the 4 of the 7 colonies had plasmids containing the right incert (sample 1,3, 7 and 8). These samples show a band at approximately 1200bp (FlhDCmut in pSB1C3 is 1248bp). the three other samples show a band heavier than 2000bp
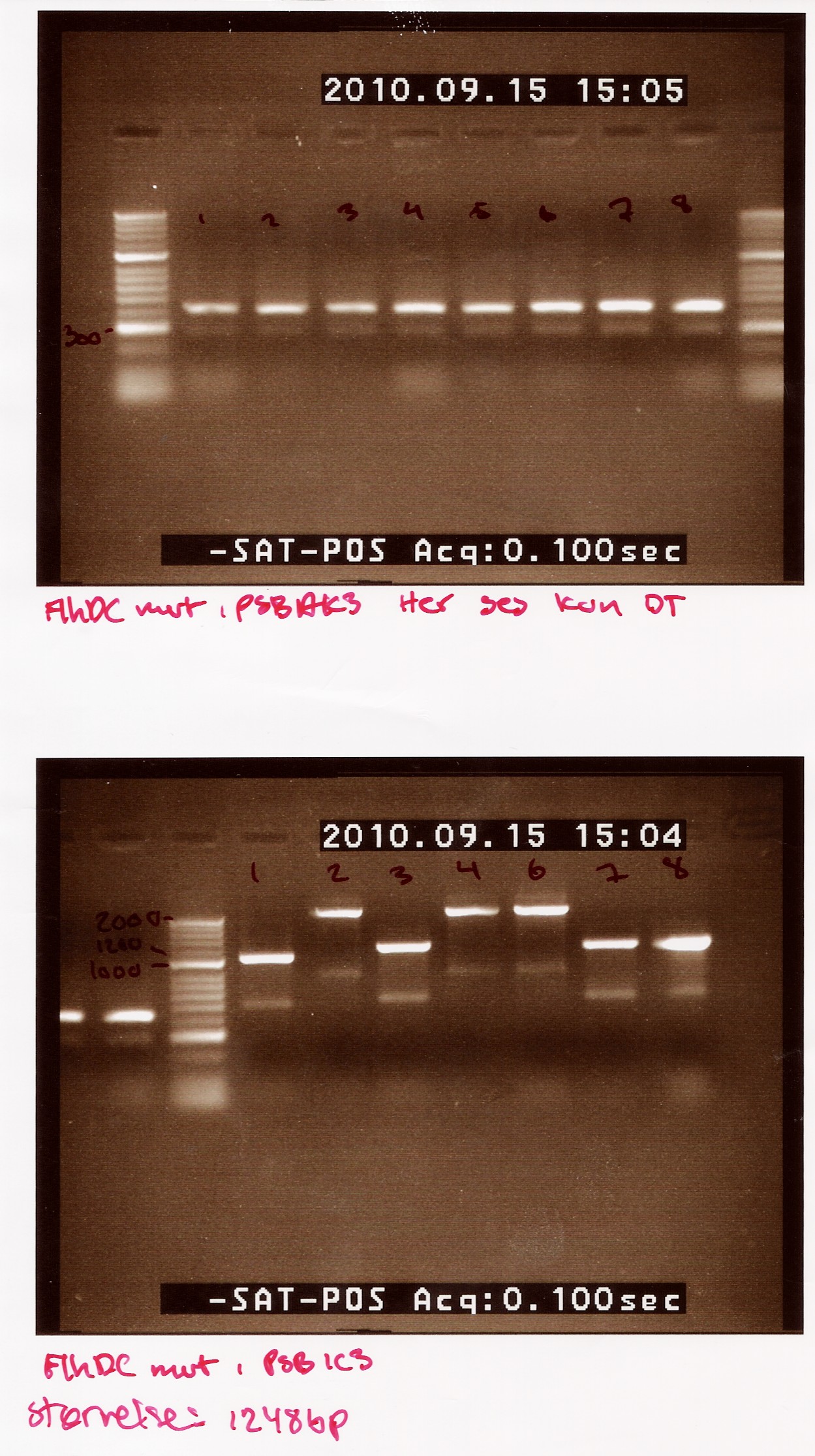
Photosensor group
Taq gradient PCR of pKJ606 with PS primers
Date: 9/7 2010
Done By: Maria and Lc
Protocol: CP1.3
Notes:
A gradient PCR was carried out to determine the appropiate annealing temperature of the new PS primers. PCR tubes were marked PSII A-H.
Premix x 9
| Taq buffer (10x) | 22.5uL |
| MgCl2 | 9uL |
| PSII fw primer | 9uL |
| PSII rv primer | 9uL |
| dNTP mix | 4.5uL |
| H20 | 158.5uL |
| Taq polymerase | 1uL |
miniprep of pKJ606 was used as template. 1uL was distrubuted into each tube.
PCR program:
| start | 95C | 2min |
| denaturating | 95C | 1min |
| annealing | se additional table | 1min |
| elongation | 72C | 2min |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
Anealing temperatures:
| 56.5C |
| 58.3C |
| 60.7C |
| 63.3C |
| 66C |
| 68.6C |
| 71C |
| 73C |
PCR product was loaded onto a 1.5 agarose gel. gene ruler DNA ladder mix (red) was used as marker.
Results:
Analysis:
Only very weak bands was observed at app. 2000 bp, and very strong bands were observed at app. 4000 bp, indicating that too much template was used in the PCR. An additional PCR was carried out using diluted miniprep as template.
--Tipi 10:27, 21 September 2010 (UTC)
Insertion of PS in pSB1C3 and pSB1AK3
Date: 9/8 - 9/12 2010
Done By: Maria and Lc
Protocol: CP1.1DE1.3RD1.1LG1.2CC1.1TR1.1CP1.3
Pfu PCR amplification of PS (no.1)
Date: 9/8 2010
Done By: Maria and Lc
Protocol:CP1.1
Notes:
5 PCR reactions are prepared. 2uL Miniprep of pKJ606 are diluted in 8uL H2O to reach a 5x dilution, and are used as template. PCR tubes are marked PSIII A-E.
Premix x6:
| pfu buffer + MgSO4 | 30uL |
| dNTP mix | 9uL |
| PSII fw primer | 9uL |
| PSII rv primer | 9uL |
| H20 | 230uL |
| pfu polymerase | 2.5uL |
| pKJ606 miniprep (5x diluted) | 10uL |
PCR program:
| start | 94C | 3min |
| denaturating | 94C | 2min |
| annealing | 68C | 30s |
| elongation | 72C | 2min30s |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
All of the PCR product was loaded onto a 1.5 agarose extraction gel. Gene ruler DNA ladder mix was used ad marker.
Results:
Analysis:
No bands were visible in the gel, indicating that the PCR reaction had been unsuccessfull. This could be due to the low elongation time, wherefore another PCR reaction with a longer elongation time was carried out.
Pfu PCR amplification of PS (no.2)
Date: 9/9 2010
Done By: Maria and Lc
Protocol:CP1.1
Notes:
5 PCR reactions are prepared. 2uL Miniprep of pKJ606 are diluted in 8uL H2O to reach a 5x dilution, and are used as template. PCR tubes are marked PSIV A-E.
Premix x6:
| pfu buffer + MgSO4 | 30uL |
| dNTP mix | 9uL |
| PSII fw primer | 9uL |
| PSII rv primer | 9uL |
| H20 | 228uL |
| pfu polymerase | 2.5uL |
| pKJ606 miniprep (5x diluted) | 12uL |
PCR program:
| start | 94C | 3min |
| denaturating | 94C | 2min |
| annealing | 68C | 30s |
| elongation | 72C | 4min20s |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
All of the PCR product was loaded onto a 1.5 agarose extraction gel. Gene ruler DNA ladder mix was used ad marker.
Results:
Analysis:
Bands were observed at app. 2000bp and bands were extracted by gel extraction
Gel extraction of PS
Date: 9/9 2010
Done By: Maria and Lc
Protocol:DE1.3
Notes:
DNA was extracted from gel according to protocol and each sample was diluted in 20uL.
Analysis:
each sample had a concentration of app. 7ng/uL. All 4 samples were pooled and used for restriction digest.
Restriction digest of PS, pSB1C3 and pSB1AK3
Date: 9/9 2010
Done By: Maria and Lc
Protocol:RD1.1DE1.3
Notes:
Restriction mixture pSB1C3:
| H2O | 24uL |
| FD green buffer | 4uL |
| EcoRI | 2uL |
| PstI | 2uL |
| pSB1C3 | 10uL |
Restriction mixture pSB1AK3:
| H2O | 24uL |
| FD green buffer | 4uL |
| EcoRI | 2uL |
| XbaI | 2uL |
| pSB1AK3 | 10uL |
Restriction mixture PS (used in pSB1C3):
| H2O | 38uL |
| FD green buffer | 8uL |
| EcoRI | 4uL |
| PstI | 4uL |
| PS | 30uL |
Restriction mixture PS (used in pSB1AK3):
| H2O | 38uL |
| FD green buffer | 8uL |
| EcoRI | 4uL |
| SpeI | 4uL |
| PS | 30uL |
Digested samples were loaded onto a 1.5% agarose extraction gel. Uncut PS, pSB1C3 and pSB1AK3 were used as controles. Gene ruler DNA ladder mix was used as marker.
Loading scheme:
| Lane | sample |
| 1 | marker |
| 2 | PS (used in pSB1C3) |
| 3 | uncut PS |
| 4 | PS (used in pSB1AK3) |
| 5 | uncut pSB1C3 |
| 6 | pSB1C3 |
| 7 | uncut pSB1AK3 |
| 8 | pSB1AK3 |
DNA was extracted from gel according to protocol.
Results:
DNA conc:
| sample | conc. (ng/uL) |
| PS (used in pSB1C3) | 5.5 |
| PS (used in pSB1AK3) | 4.2 |
| pSB1C3 | 6.3 |
| pSB1AK3 | 14.3 |
Analysis:
the purified DNA was used for ligation.
Ligation of PS and pSB1C3 and pCB1AK3
Date: 9/9 2010
Done By: Maria and Lc
Protocol:LG1.2
Notes:
For each of the ligations three ligation mixtures were prepared. vector concentrations of 10ng/uL (pSB1C3) and 15ng/uL (pSB1AK3) respectively was used for each mixture. Appropiate amount of insert was added to reac vector:insert ratios of 1:1, 1:2 and 1:4 respectively.
Ligation mixtures (PS in pSB1C3):
| L1 | L2 | L3 | |
| T4 ligase buffer | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB1C3 | 1.5uL | 1.5uL | 1.5uL |
| PS | 1.5uL | 3uL | 6uL |
| H20 | 14uL | 12.5uL | 9.5uL |
Ligation mixtures (PS in pSB1AK3):
| L1 | L2 | L3 | |
| T4 ligase buffer | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB1C3 | 1uL | 1uL | 1uL |
| PS | 2uL | 4uL | 8uL |
| H20 | 14uL | 12uL | 8uL |
The samples was incubated at 17C ON at used for transformation
Transfomation of ligated plasmin in Top 10 E.coli
Date: 9/10 2010
Done By: Maria and Lc
Protocol:CC1.1TR1.1
Notes:
The compotent cells and transformation was carried out according to protocol. Cells transformed with ligations of PS in pSB1AK3 was plated on kanamycine plates.
Results:
the controle plates were okay, and there was many colonies on plates with cells transformed with ligations of pSB1C3 and PS. There was 10-20 colonies on the plates with cells transformed with ligation of PS and pSB1AK3.
Analysis:
The transformation was successfull and colonies was selected and used in coloni PCR.
--Tipi 13:41, 26 September 2010 (UTC)
Colony PCR of transformation of PS in pSB1C3 and pSB1AK3 in TOP10 E.Coli
Date: 09/11
Done by: LC & Maria
Methods: Colony PCR
Protocols: CP1.3[1]
Notes:
Premix:
45 µl 10xTAQ Buffer
18 µl MgCl2
18 µl VF2
18 µl VR
9 µl dNTP
63 µl H2O
2,25 µl TAQ Polymerase
The chosen colonies were lysed in 15 ul of H20.
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
Results: For the ligation into pSB1C3, colonies B, F and H showed bands at the right length. For the other reatcion (in pSB1AK3), colonies A to D showed bands with the correct length. The colonies with bands at the right length will be miniprepped and sent for sequencing.

Control restriction digest of cPCR product of PS in pSB1C3 and pSB1AK3
Date: 09/12
Done by: LC & Maria
Methods: Restriction digest
Protocols: RD1.1[2]
Notes:
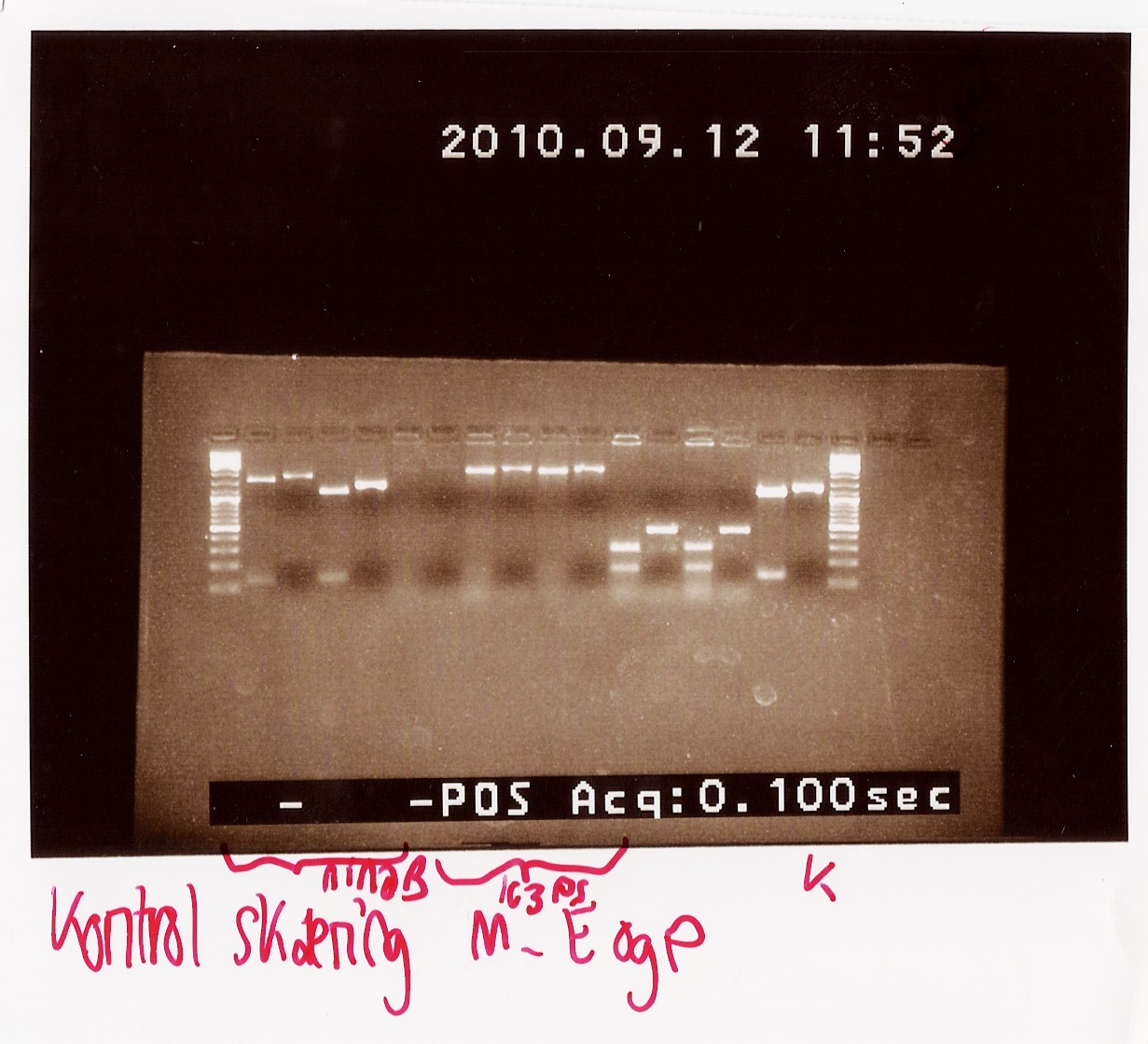
The afore mentioned samples (B, F and H of PS in pSB1C3 and A and D in pSB1AK3) were cut with EcoRI and PstI to see if it was 1) possible to cut them at all, thereby confirming the presence of biobrick prefix and suffix and 2) determining if the insert in the plasmid has the right length.
Mix for 1 RD reaction:
12 ul H2O
1 ul PstI
1 ul EcoRI
2 ul Fast digest green buffer
5 ul PCR product
Uncut PCR product and RFP were also loaded on the gel as a control.
Results: All samples pSB1C3 samples were cut as expected (to around 2000 BP), but the restriction digest of pSB1AK3 sample A and D failed, so that one will have to be repeated with sample B and C, as these only showed a cut double terminator without the PS insert.
Second control restriction digest of cPCR product of PS pSB1AK3
Date: 09/12
Done by: LC & Maria
Methods: Restriction digest
Protocols: RD1.1[3]
Since samples A and D fromt he colony PCR gave inadequate results when cut with EcoRI and PstI, we had to repeat the restriction digest with the remaining two samples that were the right length on the cPCR, sample B and C. Samples E, F and G were also cut to confirm that there were no errors in naming the samples that resulted in sample A and D not giving the expected results.
Restriction mixture:
72 ul H2O
6 ul PstI
6 ul EcoRI
12 ul Fast digest green buffer
5 ul PCR product per sample (x6)
Uncut PCR product was loaded on the gel as a control.
Results: Only sample C showed bands at the correct length (around 2180 BP), where as all other bands were at the length of a double terminator (b0015) without the photosensor insert. All samples were properly cut indicating the presence of biobrick pre- and suffix in these. Sample C will be miniprepped and sent off to sequencing.
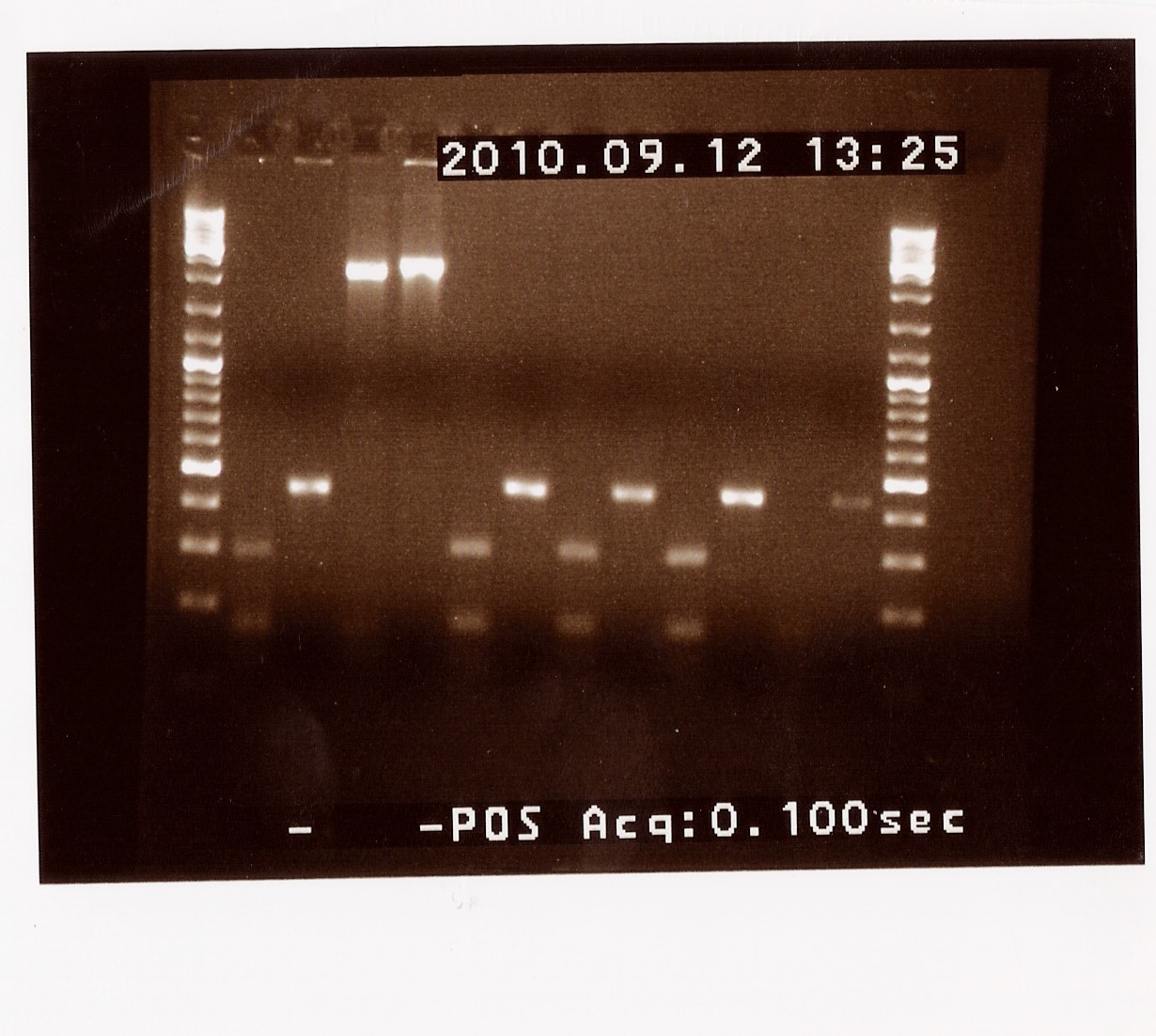
Second round of colony PCR on PS ligated into pSB1AK3 (containing double terminator
Date: 09/12
Done by: LC & Maria
Methods: Colony PCR
Protocols: CP1.3[4]
Notes:
Since only sample C from the previous colony PCR looked right after a restriction digest we decided to run another round of colony PCR on the photosensor ligated into pSB1AK3, to find more colonys containing the right (ligated!) plamid.
The original samples A - D were also used as a template, now called I, J, K and L. Sample M to Y were new colonies.
Premix:
42,5 µl 10xTAQ Buffer
17 µl MgCl2
17 µl VF2
17 µl VR
8,5 µl dNTP
59,5 µl H2O
2,125 µl TAQ Polymerase
The chosen colonies were lysed in 15 ul of H20.
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
Results: Samples K (former C) and Q produced bands at the right length of around 2150 BP, which eans that sample Q will also be miniprepped and send to sequencing together with the previously chosen sample C (now K).
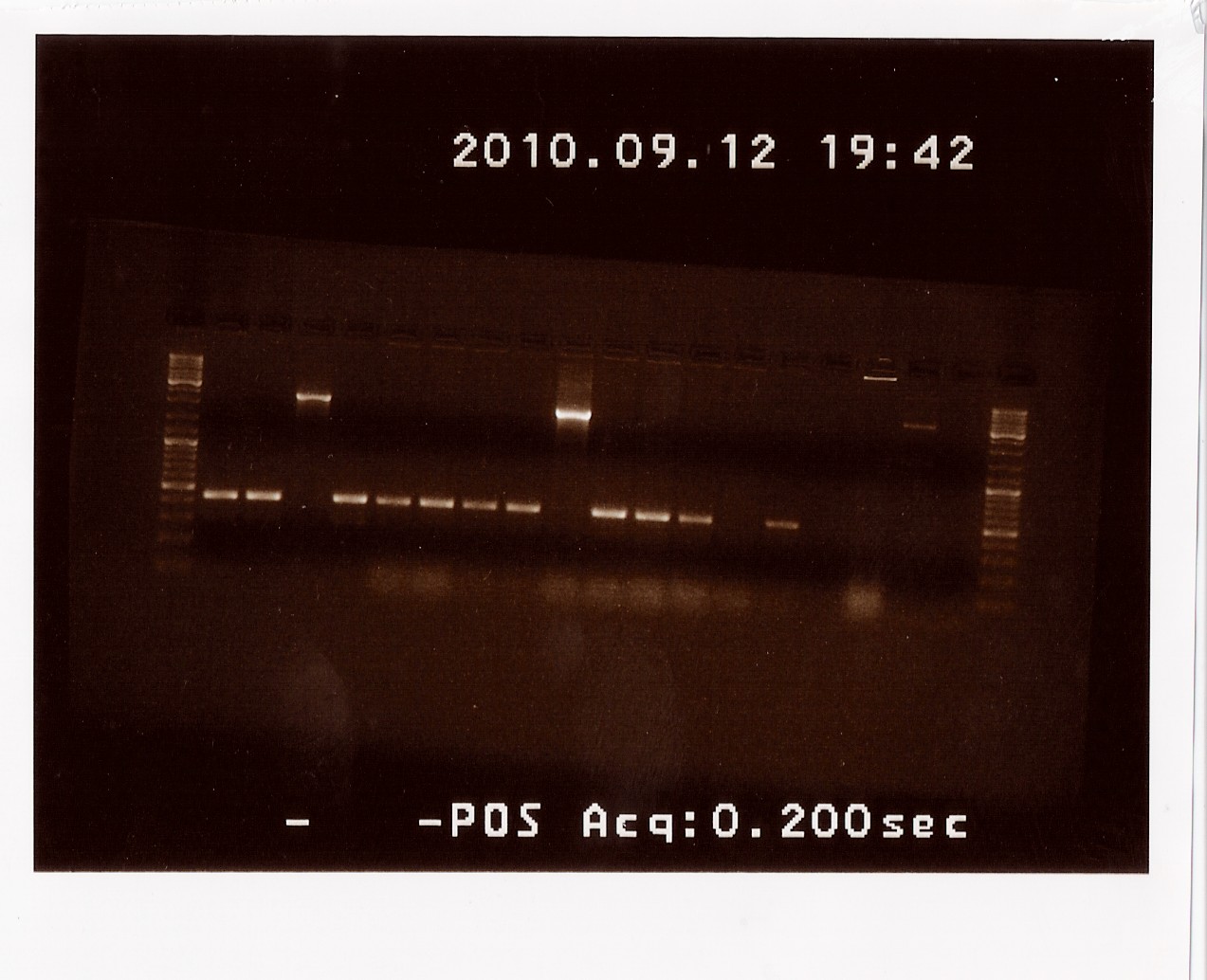
Miniprep and controle digestion of PS in pSB1C3 and pSB1AK3
Date: 9/13 2010
Done By: Maria and Lc
Protocol:MP1.2RD1.1
Notes:
Coloni B, F and H of PS in pSB1C3 and coloni C of PS + double terminator in pSB1AK3 was miniprep'ed according to prtocol. To ensure that the plasmids contains the insert as expected, the miniprep is cut with EcoRI and PstI.
the 2 minipreps of each coloni are diluted in 20uL and 50uL respectively.
Restriction mixture x4:
| H2O | 48uL |
| FD green buffer | 8uL |
| EcoRI | 4uL |
| PstI | 4uL |
| Plasmid | 4x5uL |
The digested products are loaded onto a 1% agarose gel. 2.5uL of undigested plasmid are used as controle. Gene ruler DNA ladder mix are used as marker
Loading scheme:
| Lane | Sample |
| 1 | marker |
| 2 | digested plasmid from col. B |
| 3 | undigested plasmid from col. B |
| 4 | digested plasmid from col. F |
| 5 | undigested plasmid from col. F |
| 6 | digested plasmid from col. H |
| 7 | undigested plasmid from col. H |
| 8 | digested plasmid from col. C |
| 9 | undigested plasmid from col. C |
| 10 | marker |
concentrations of miniprep was measured on nanodrop.
Results:
nanodrop:
| sample | conc. ng/uL | 260/280 | 260/230 |
| PS i pSB1C3 B1 | 232.86 | 1.91 | 2.17 |
| PS i pSB1C3 B2 | 134.5 | 1.93 | 2.17 |
| PS i pSB1C3 F1 | 229.32 | 1.9 | 2.18 |
| PS i pSB1C3 F2 | 144.11 | 1.92 | 2.15 |
| PS i pSB1C3 H1 | 190.88 | 1.91 | 2.2 |
| PS i pSB1C3 H2 | 137.54 | 1.89 | 2.15 |
| PS + double terminator in pSB1AK3 C1 | 227.6 | 1.91 | 2.2 |
| PS + double terminator in pSB1AK3 C2 | 138.57 | 1.92 | 2.18 |
Analysis:
In the lanes containing the digested pSB1C3 plasmids bands at app. 2000bp are observed corresponding to PS. The lane containing digested pSB1AK3 plasmids shows a band at app. 2100bp corresponding to the size of PS and the double terminator. Miniprep of coloni B, F and C are sent to sequentation.
--Tipi 14:44, 26 September 2010 (UTC)
Miniprep of PS+B0015 sample Q and pSB3T5 containing J13002
Date: 27/7
Methods: ON, Miniprep
Protocol: MP1.1
Experiment done by: LC & Maria
Notes: PS+B0015 sample Q (1) was eluded in 20 ul to get a concentration over 200 ng/ul, so we would directly be able to send it in for sequencing. All other samples were eluded in 50 ul of elution buffer.
Results: The concentrations were as followed:
PS+B0015 (1): 180 ng/ul
PS+B0015 (2): 130 ng/ul
pSB3T5+J13002 (1 and 2): 50 ng/ul
Sample Q did not fulfill DNA technologies sample specifications for sequencing we waited with sending it in, since we were expecting the results from sample C which was sent in earlier. Since those returned positive, sample Q was never sent in, since we just proceeded with sample C.
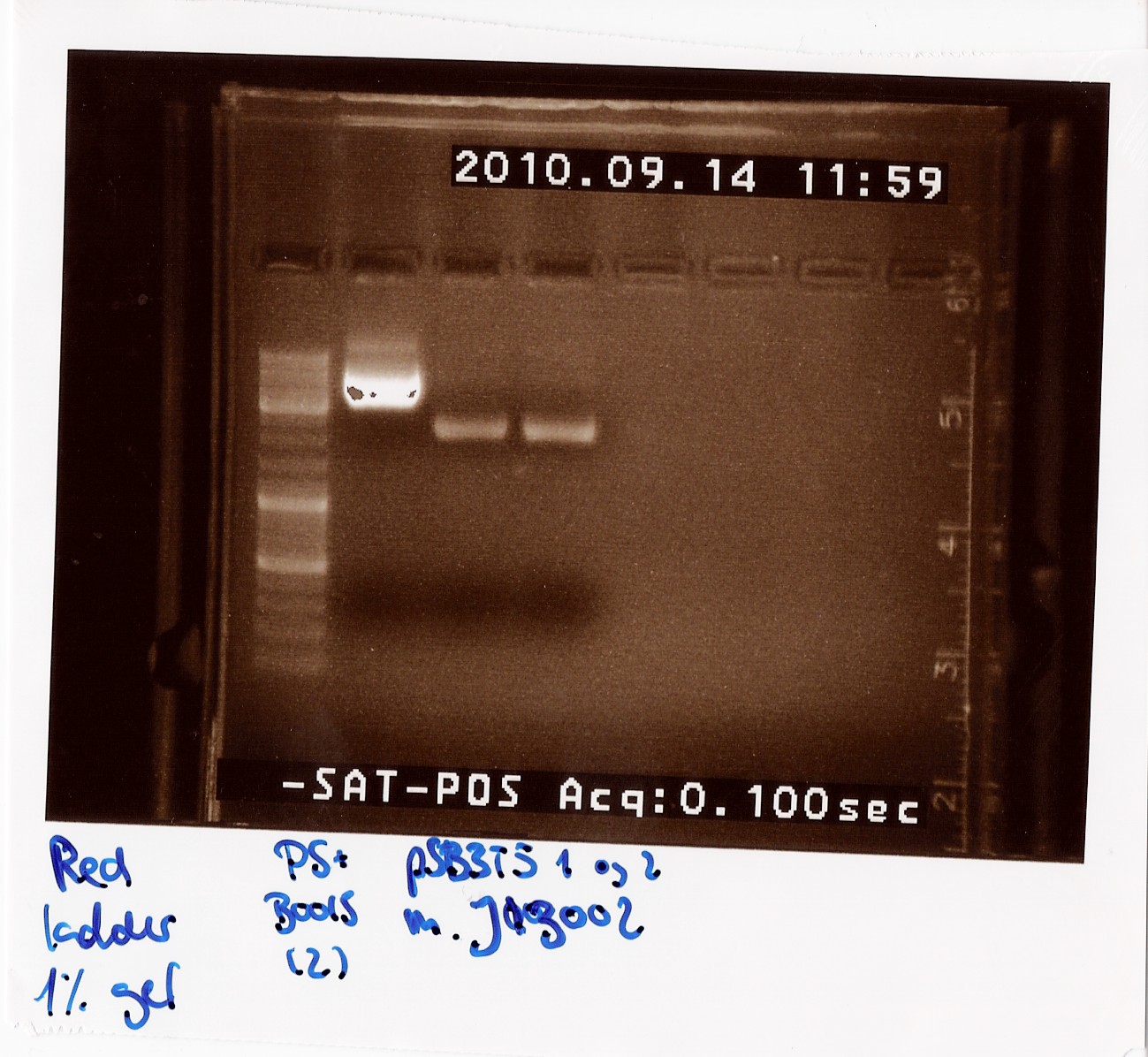
 "
"