Team:Lethbridge/Notebook/Lab Work/June
From 2010.igem.org
Adam.smith4 (Talk | contribs) (→June 10/2010) |
Adam.smith4 (Talk | contribs) (→June 2010) |
||
| Line 423: | Line 423: | ||
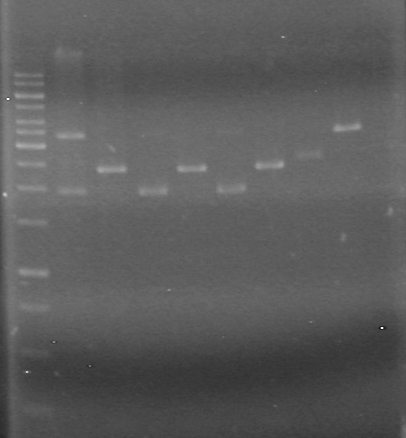
[[image:100608-AV.AS.HS(3).jpg|200px|none]]<br> | [[image:100608-AV.AS.HS(3).jpg|200px|none]]<br> | ||
Looks like the mms6 DNA was not cut at all. Therefore is doubtful that the ligations will work.<br><br> | Looks like the mms6 DNA was not cut at all. Therefore is doubtful that the ligations will work.<br><br> | ||
| + | |||
| + | ===June 8/2010 - Evening=== | ||
| + | <b>Objective:</b> Transform pLacI-sRBS-Lum-dT constructs assembled via three antibiotic assembly on June 3/2010. Also transform mms6-dT constructs assembled today using old insertion method.<br> | ||
| + | <b>Method:</b> Follow [[Team:Lethbridge/Notebook/Protocols|transformation protocol]]. | ||
| + | <b>Results:</b> No colonies anywhere<br> | ||
===June 9/2010=== | ===June 9/2010=== | ||
Revision as of 02:49, 12 June 2010
Contents |
June 2010
June 1/2010
JV quantified the amount of DNA in gels run to date using ImageJ software. Results to be posted in working plasmids box.
Objective: Transform plasmids into DH5α
Method: Follow competent cell transformation protocol to transform the following:
From our ligations:
- pLacI-sRBS-Lumazine-dT
- pLacI-sRBS-Lumazine-dT
- mms6 (A6)
- mms6 (B6)
- xylE (C4)
- xylE (B4)
From the 2010 Parts Distribution:
- ECFP (Bba_E0020)
- EYFP (Bba_E0030)
- BglII Endonuclease (Bba_K112106)
June 2/2010
(In Lab: JV)
Objective: Isolate plasmid DNA of RBS-xylE (BBa_J33204) from DH5α cells and confirm results.
Method: "Mini-prep" the plasmid DNA using boiling lysis miniprep. Then restrict the DNA once and run on a 1% agarose gel (TAE).
Restriction Reaction
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 15.75 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
| EcoRI | 0.25 |
Unrestricted Control
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 16 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
DNA was restricted for 80 minutes at 37oC.
Analyzed results on a 1% agarose gel. Load order as follows:
| Lane | Sample | Volume Sample (µL) | Volume Loading Dye (µL) |
| 1 | Restricted RBS-xylE | 10 | 2 |
| 1 | Unestricted RBS-xylE† | 1 | 2 |
| 1 | 1kb Ladder†† | 2 | 2 |
† Added 9µL MilliQ H2O
†† Added 8µL MilliQ H2O
Ran gel at 100V from 2 hours.
Results:
Conclusions: Plasmid DNA prep and restriction was successful.
Objective: Ligate rbs-xylE (Bba_J33204) to our double terminator, and insert it into the pSB1T3 plasmid backbone.
Method:
- Restrictions
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
- Restrict the double terminator with XbaI and PstI (Tango Buffer)
- Restrict pSB1T3 with EcoRI and PstI (Red Buffer)
Component Volume (µL) MilliQ H2O 15.5 Buffer 2 pDNA 2 Enzyme 0.25 + 0.25 Set up control reaction as follows:
- MilliQ H2O - 16µL
- Buffer - 2µL
- pDNA - 2µL
Incubated reactions for 65 minutes at 37oC
Killed enzymes by incubating reactions for 10 minutes at 65oC
- Ligation
Reaction set up as follows:- T4 DNA ligase - 0.25µL
- rbs-xylE - 5µL
- dT - 3µL
- pSB1T3 - 8µL
- 10x Ligation Buffer - 2µL
- MilliQ H2O - 1.75µL
Killed enzymes by incubating reactions for 10 minutes at 80oC</ul>June 2/2010 - Evening
Objective: Set up new ligations of pLacI and sRBS-Lum-dT according to Tom Knight's protocol. Previous ligation had very little DNA.
Relevant Information:
- Want a final mass of 25ng of each pDNA in the ligation mix.
- Final concentration of pDNA in restriction digest should be 25-50ng/µL.
- Tom Knight's restriction reaction is 50µL, therefore there should be 1000ng pDNA in each restriction digest.
- Identified the following plasmids in our working plasmids box:
Common Name Location Concentration (ng/µL) Volume/rxn (µL) pLacI Maxiprep A9 990 ~1 pLacI (B1) A6 440 ~2 sRBS-Lum-dT (2) A1 965 ~1 sRBS-Lum-dT (1) A2 1145 ~1 sRBS-Lum-dT Maxiprep B8 4780 ~.2 sRBS-Lum-dT B7 4375 ~.25 sRBS-Lum-dT (1) G2 335 ~3 sRBS-Lum-dT (2) G3 965 ~2 - Make a 1:10 dilution of sRBS-Lum-dt maxiprep (D8) and sRBS-Lum-dT (B7). 0.5µL pDNA in 4.5µL water.
- Cut pLacI with EcoRI and SpeI
- Cut sRBS-Lum-dT with XbaI and PstI
- Cut pSB1T3 with EcoRI and PstI
- Will have total of 12 ligation reactions, want 12x2µL of pSB1T3 to add to each, therefore want 25µL of pSB1T3.
Method:
Restriction
Name [pDNA] (ng/µL) Volume
pDNA (µL)Volume
Water (µL)Volume
Buffer (µL)Enzymes Total Volume sRBS-Lum-dT (A1) 965 1 43.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (A2) 1145 1 43.5 5 0.25µL XbaI
0.25µL PstI50 pLacI Maxiprep (A1) 990 1 43.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT Maxiprep(B8) 4780 2 (of 1:10 dilution) 42.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (B7) 4375 2.5 (of 1:10 dilution) 42 5 0.25µL XbaI
0.25µL PstI50 pLacI (D6) 440 2 42.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT (G2) 335 3 41.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (G3) 540 2 42.5 5 0.25µL XbaI
0.25µL PstI50 pSB1T3 25 12.5 7 5 0.25µL EcoRI
0.25µL PstI50 Incubate for 30 minutes at 37oC (Start- 12:10pm; End- 12:40pm)
Heat kill enzymes at 80oC for 20 minutes
Ligation:
In a 10µL final volume, add:- 2µL of sRBS-Lum-dT component
- 2µL of pLacI component
- 2µL of pSB1T3 component
- 1µL of T4 Buffer
- 0.25µL of T4 DNA Ligase
- 2.75µL of MilliQ H2O
Incubate for 30 minutes at room temperature to ligate
Incubate for 20 minutes at 80oC to heat kill
June 3/2010
Carried out protocol described in June 2/2010 - Evening
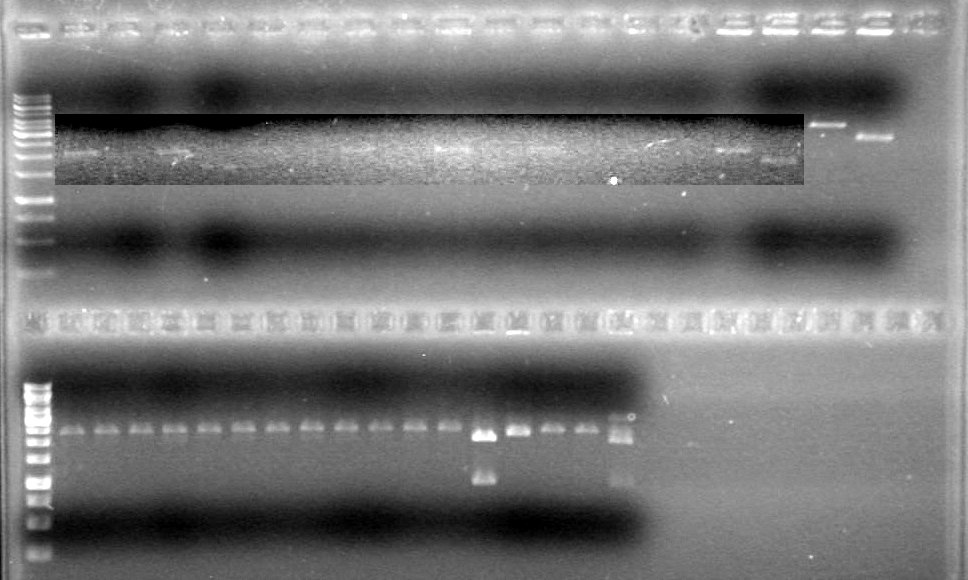
Analyzed results on 1% agarose gel.Load order as follows:
Lane Gel 1
SampleGel 1 Load Gel 2
SampleGel 2 Load 1 1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O2 Restricted
sRBS-Lum-dT (A1)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(A1)10µL DNA
2µL Dye3 Unrestricted
sRBS-Lum-dT (A1)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(A1)10µL DNA
2µL Dye4 Restricted
sRBS-Lum-dT (A2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(A2)10µL DNA
2µL Dye5 Unrestricted
sRBS-Lum-dT (A2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(A2)10µL DNA
2µL Dye6 Restricted
sRBS-Lum-dT (B8)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(B7)10µL DNA
2µL Dye7 Unrestricted
sRBS-Lum-dT (B8)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(B7)10µL DNA
2µL Dye8 Restricted
sRBS-Lum-dT (B7)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(G2)10µL DNA
2µL Dye9 Unrestricted
sRBS-Lum-dT (B7)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(G2)10µL DNA
2µL Dye10 Restricted
sRBS-Lum-dT (G2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(G3)10µL DNA
2µL Dye11 Unrestricted
sRBS-Lum-dT (G2)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(G3)10µL DNA
2µL Dye12 Restricted
sRBS-Lum-dT (G3)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(D6)+sRBS-Lum-dT(B8)10µL DNA
2µL Dye13 Unrestricted
sRBS-Lum-dT (G3)10µL DNA
2µL DyepSB1T3 Ligation of:
pLacI(A9)+sRBS-Lum-dT(B8)10µL DNA
2µL Dye14 Restricted
pLacI (A9)10µL DNA
2µL DyeRestricted
rbs-xylE10µL DNA
2µL Dye15 Unrestricted
pLacI (A9)10µL DNA
2µL DyeUnrestricted
rbs-xylE10µL DNA
2µL Dye16 Restricted
pLacI B1 (D6)10µL DNA
2µL DyeRestricted
pSB1T310µL DNA
2µL Dye17 Unrestricted
pLacI B1 (D6)10µL DNA
2µL DyeUnrestricted pSB1T3 10µL DNA
2µL Dye18 Unrestricted
dT10µL DNA
2µL DyepSB1T3 Ligation of:
rbs-xylE+dT10µL DNA
2µL Dye19 Restricted
dT10µL DNA
2µL DyeRan gel at 100V for 90 minutes.
Results:
Conclusions:
June 3/2010 - Evening
Objective: Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel.
Method:
Ligation:
In a 10µL final volume, add:- 2µL of sRBS-Lum-dT component
- 2µL of pLacI component
- 2µL of pSB1T3 component
- 1µL of T4 Buffer
- 0.25µL of T4 DNA Ligase
- 2.75µL of MilliQ H2O
Incubate for 30 minutes at room temperature to ligate
Incubate for 20 minutes at 80oC to heat kill
Following ligation, transformed using transformation protocol. Plates incubated in 37oC incubator for 44 hours.
Results: Only plate pLacI (D6) + sRBS-Lum-dT (G2) + pSB1T3 grew; had 2 colonies. Control plate did not grow, acidentally plated on tetracycline plate instead of ampicillin (pUC19).
Follow-up: Inoculated 5mL LB media (tetracycline positive) with cells from the transformation plates and incubated at 37oC overnight. (June 5/2010).
Objective: Ligate pBad-TetR part with fluorescent protein part in pSB1C3 backbone.
Method:
Restriction
Name Volume
pDNA (µL)Volume
Water (µL)Volume
Buffer (µL)Enzymes Total Volume pSB-NEYFP (B4) .8 43.7 5 0.25µL XbaI
0.25µL PstI50 pSB-CEYFP (B5) .9 43.6 5 0.25µL XbaI
0.25µL PstI50 pBad-TetR) .3 44.2 5 0.25µL EcoRI
0.25µL SpeI50 NEYFP (E1) 4.3 40.7 5 0.25µL XbaI
0.25µL PstI50 NEYFP (E2) 0.3 42.2 5 0.25µL XbaI
0.25µL PstI50 Fusion CEYFP (E3) 3.9 40.6 5 0.25µL XbaI
0.25µL PstI50 Fusion CEYFP (E4) 2.0 42.5 5 0.25µL XbaI
0.25µL PstI50 Fusion CEYFP (E5) 3.0 41.5 5 0.25µL XbaI
0.25µL PstI50 CEYFP (E6) 0.6 43.9 5 0.25µL XbaI
0.25µL PstI50 CEYFP (E7) 0.5 44.0 5 0.25µL XbaI
0.25µL PstI50 pBad-TetR (F4) 2.5 42 5 0.25µL EcoRI
0.25µL SpeI50 pBad-TetR (F5) 1.7 42.8 5 0.25µL EcoRI
0.25µL SpeI50 pSB-CEYFP (G4) 2.9 41.6 5 0.25µL XbaI
0.25µL PstI50 pSB1C3 15.5 46 0.25µL EcoRI
0.25µL PstI62 Incubated at 37oC for 75 minutes.
- Used Red buffer for the EcoRI/SpeI and EcoRI/PstI digests
- Used Tango buffer for the XbaI/PstI digests
- Did not heat kill upon removal from incubation, put directly into -20oC fridge.
Continue Ligation on Saturday (See below).
June 5/2010
(In the lab:AS)
Objective: Ligate restriction products from June 3/2010.
Relevant information:
- Have 3 tubes of part 1 (pBad-TetR)
- In ampicillin backbone
- Have 10 tubes of part 2 (fluorescent protein - various)
- In ampicillin backbone
- Will have 30 combinations
- Will use pSB1C3 as plasmid backbone
- Used most of the pSB1T3 and want to save remainder for creating new backbone via PCR.
Method:
*Restriction digests were not heat killed after reactions. Freezing probably killed the restriction enzymes, but I will hea kill them at 80oC for 20 minutes anyways prior to adding Ligase.- Cool on ice for 10 minutes before adding ligase.
Master Mix Volume/tube (µL) Total Volume (µL) DNA 6 --- 10x Buffer 1 32 T4 DNA Ligase .25 8 MilliQ H2O 2.75 88 - Add 4µL master mix to each DNA tube.
Follow-up: Ligation reactions will be transformed into DH5α cells
June 6/2010
(In Lab: JV, HS)
Objective:
Isolate the following plasmid DNA from DH5α:
- pLacI-sRBS-Lumazine-dT in pSB1T3 (colony 2)
- pLacI-sRBS-Lumazine-dT in pSB1T3 (colony 1)
Method:
Followed boiling lysis miniprep protocol. Eluted with 10µL Milli-Q H2O and RNase A.
Notes:
- Placed colony 2 in cell E10 of glycerol stocks and J6 of working plasmid box.
- Placed colony 2 in cell F1 of glycerol stocks and J5 of working plasmid box.
Objective:
Transformed the following plasmid DNA into DH5α cells:
- pBad-TetR-CEYFP: (F5+E6), (B10+E7), (B10+E6), (F4+E6), (F5+E4), (F4+E7)
- pBad-TetR-Fusion CEYFP: (F5+E3), (B10+E4), (F4+E3), (B10+E3), (B10+E5), (F4+E4), (F5+E4), (F5+E5), (F4+E5)
- pBad-TetR-pSB CEYFP: (F4+B5), (B10+G4), (F4+G4), (F4+B5), (F5+B5), (F5+G4)
- pBad-TetR-NEYFP: (F5+E1), (B10+E1), (F4+E2), (F5+E2), (B10+E2), (F4+E1)
- pBad-TetR-pSB NEYFP: (F5+B4), (F4+B4), (B10+B4)
- Positive control -> DH5α + pSB1C3
- Negative control -> DH%α + Milli-Q H2O
Method:
Followed Competent Cell Transformation protocol and used chloramphenicol as an antibiotic. We plated all 200µL of DNA onto the plates. The plates were incubated at 37oC from 4:30pm to 10:00am.Results:
None of the plates showed any growth.June 8/2010
(In the lab: JV)
Objective: Follow the overexpression of our pLacI-sRBS-Lum-dT construct.
Method: FILL ME OUT!!!!!!
Results:Time OD600 F1 OD600 E10 0 0.118 0.103 30 0.133 0.111 30 0.145 0.116 90 120 0.160 0.124 150 0.120 0.093 180 0.129 0.100 210 0.145 0.122 240 0.158 0.145 270 0.171 0.178 300 0.194 0.222 330 0.223 0.280 360 0.252 0.364 390 0.296 0.458 420 0.338 0.557† 450 0.394 0.656 480 0.453 0.675 510 0.530 0.688 540 0.598† 0.706 600 0.633 0.752 660 0.653 720 0.679 ∞ 1.278 † Overexpression induced by adding 1mM IPTG.
Following overexpression, 1mL of cells was removed from the culture, spun down at ~13000xg for 20 seconds, excess media removed and rinsed with water.
Suspended cells in 8M urea, mixed with 6x dye and ran on 18% SDS-PAGE gel for 90 minutes at 200V. Gel stained overnight.
Results: IMAGE TO COME!!!!
Objective: Calculate quantity of DNA in pBad-TetR and fluorescent protein mini-preps by staining an agarose gel.
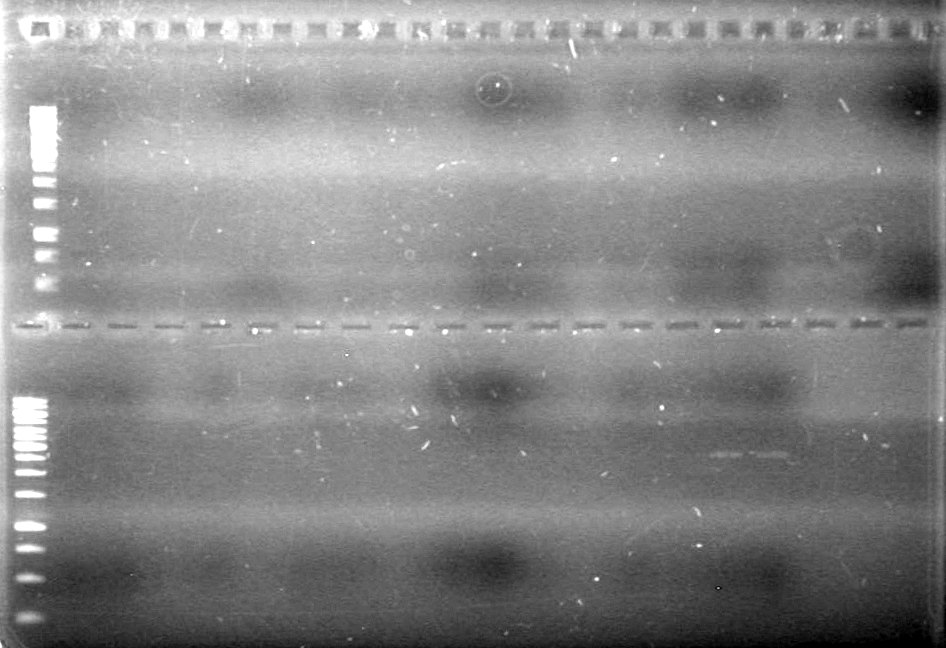
Method: Restrict plasmid DNA (done by AV,HB,TF on June 7/2010) and run on a 1% TAE agarose gel (JV).
Lane Gel 1
SampleGel 1 Load Gel 2
SampleGel 2 Load 1 1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O1kb Ladder 2µL dye, 2µL ladder
8µL MilliQ H2O2 Restricted
EYFP (B1)10µL DNA
2µL DyeRestricted
Fusion CEYFP (E3)10µL DNA
2µL Dye3 Unrestricted
EYFP (B1)10µL DNA
2µL DyeUnrestricted
Fusion CEYFP (E3)10µL DNA
2µL Dye4 Restricted
pSB-CEYFP (B5)10µL DNA
2µL DyeRestricted
Fusion CEYFP (E4)10µL DNA
2µL Dye5 Unrestricted
pSB-CEYFP (B5)10µL DNA
2µL DyeUnrestricted
Fusion CEYFP (E4)10µL DNA
2µL Dye6 Restricted
ECFP (F2)10µL DNA
2µL DyeRestricted
EYFP (E10)10µL DNA
2µL Dye7 Unrestricted
ECFP (F2)10µL DNA
2µL DyeUnrestricted
EYFP (E10)10µL DNA
2µL Dye8 Restricted
pSB-CEYFP (G4)10µL DNA
2µL DyeRestricted
pSB NEYFP (B4)10µL DNA
2µL Dye9 Unrestricted
pSB-CEYFP (G4)10µL DNA
2µL DyeUnrestricted
pSB NEYFP (B4)10µL DNA
2µL Dye10 Restricted
EYFP (G1)10µL DNA
2µL DyeRestricted
ECFP (F3)10µL DNA
2µL Dye11 Unrestricted
EYFP (G1)10µL DNA
2µL DyeUnrestricted
ECFP (F3)10µL DNA
2µL Dye12 Restricted
NEYFP (E2)10µL DNA
2µL DyepBad-TetR (F5) 10µL DNA
2µL Dye13 Unrestricted
NEYFP (E2)10µL DNA
2µL DyeRestricted
Fusion CEYFP (E5)10µL DNA
2µL Dye14 Restricted
pBad-TetR (B10)10µL DNA
2µL DyeUnrestricted
Fusion CEYFP (E5)10µL DNA
2µL Dye15 Unrestricted
pBad-TetR (B10)10µL DNA
2µL DyepBad-TetR (F4) 10µL DNA
2µL Dye16 Restricted
CEYFP (E6)10µL DNA
2µL DyeRestricted
pSB1T310µL DNA
2µL Dye17 Unrestricted
CEYFP (E6)10µL DNA
2µL DyeUnrestricted pSB1T3 10µL DNA
2µL Dye18 Restricted
NEYFP (E1)10µL DNA
2µL Dye10µL DNA
2µL Dye19 Unrestricted
NEYFP (E1)10µL DNA
2µL Dye20 Restricted
ECFP (F1)10µL DNA
2µL Dye21 Unrestricted
ECFP (F1)10µL DNA
2µL Dye22 Restricted
EYFP (E9)10µL DNA
2µL Dye23 Unrestricted
EYFP (E9)10µL DNA
2µL Dye24 Restricted
CEYFP (E7)10µL DNA
2µL Dye25 Unrestricted
CEYFP (E7)10µL DNA
2µL Dye26 Restricted
EYFP (E8)10µL DNA
2µL Dye27 Unrestricted
EYFP (E8)10µL DNA
2µL DyeRan gel at 100V for 90 minutes.
Results:
No bands visible except for pSB1T3 lanes, therefore could not quantify anything that had not already been quantified.
Also, purification of DNA done on June 7/2010 seemed to reduce amount of pDNA in sample.
Objective: Restrict mms6 (D9,D10) and dT (C1) so we can ligate the dT onto the mms6 coding region.
Method:
mms6 Restriction
- 2µL mms6 pDNA
- 2µL Red buffer
- 0.25µL EcoRI
- 0.25µL SpeI
- 15.5µL MilliQ H2O
dT Restriction
- 2µL dT pDNA
- 2µL Orange buffer
- 0.25µL EcoRI
- 0.25µL XbaI
- 15.5µL MilliQ H2O
Incubated for 1 hour at 37oC
Heat shock on heat block (80oC) for 20 minutes
Ligation- 2µL dT pDNA
- 2µL mms6 pDNA
- 0.25µL T4 DNA Ligase
- 1µL 10x buffer
- 4.75µL MilliQ H2O
Analyze results on 1% TAE agarose gel
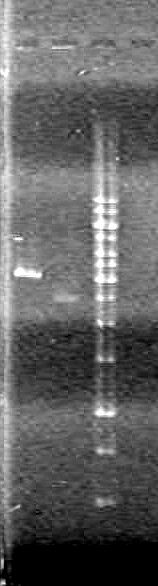
Lane Sample Load (µL) 1 1kb ladder 0.5 ladder; 2 dye; 9.5 MilliQ H2O 2 Unrestricted mms6 (D9) 10 DNA; 2 Dye 3 Restricted mms6 (D9) 10 DNA; 2 Dye 4 Unrestricted mms6 (D10) 10 DNA; 2 Dye 5 Restricted mms6 (D10) 10 DNA; 2 Dye 6 Unrestricted dT (C1) 10 DNA; 2 Dye 7 Restricted dT (C1) 10 DNA; 2 Dye 8 Empty 9 Empty 10 Empty Results:
Looks like the mms6 DNA was not cut at all. Therefore is doubtful that the ligations will work.
June 8/2010 - Evening
Objective: Transform pLacI-sRBS-Lum-dT constructs assembled via three antibiotic assembly on June 3/2010. Also transform mms6-dT constructs assembled today using old insertion method.
Method: Follow transformation protocol. Results: No colonies anywhere
June 9/2010
(in the lab: TF, JV)
Objective: Transform mms6-dT ligation reactions from June 8/2010.
Method: Followed transformation protocol and transformed the following:- mms6 (D9) + dT (C1)
- mms6 (D10) + dT (C1)
- pUC19 (positive control)
- Water (negative control)
Results: No transformants on plates.
June 10/2010
(In the lab: AV, HB, JV)
Objective: Repeat ligation of mms6 (B9,D9,D10) and dT (C1).
Method:
mms6 Restriction
- 2µL mms6 pDNA
- 2µL Red buffer
- 0.25µL EcoRI
- 0.25µL SpeI
- 15.5µL MilliQ H2O
dT Restriction
- 2µL dT pDNA
- 2µL Orange buffer
- 0.25µL EcoRI
- 0.25µL XbaI
- 15.5µL MilliQ H2O
Incubated for 1 hour at 37oC.
Killed enzymes by heating to 80oC for 20 minutes
Ligation- 5µL dT pDNA
- 5µL mms6 pDNA
- 0.5µL T4 DNA Ligase
- 2µL 10x buffer
- 7.5µL MilliQ H2O
Incubated at room temperature overnight.
Results:
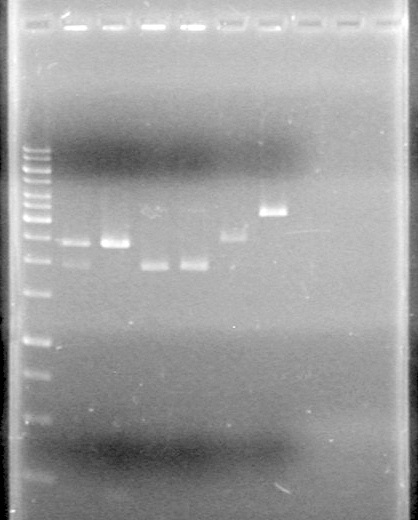
Analyzed on 1% TAE agarose gel:Lane Sample Load (µL) 1 1kb ladder 0.5 ladder; 2 dye; 9.5 MilliQ H2O 2 Unrestricted mms6 (B9) 5 DNA; 2 dye; 5 MilliQ H2O 3 Restricted mms6 (B9) 5 DNA; 2 dye; 5 MilliQ H2O 4 Unrestricted mms6 (D9) 5 DNA; 2 dye; 5 MilliQ H2O 5 Restricted mms6 (D9) 5 DNA; 2 dye; 5 MilliQ H2O 6 Unrestricted mms6 (D10) 5 DNA; 2 dye; 5 MilliQ H2O 7 Restricted mms6 (D10) 5 DNA; 2 dye; 5 MilliQ H2O 8 Unrestricted dT (C1) 5 DNA; 2 dye; 5 MilliQ H2O 9 Restricted dT (C1) 5 DNA; 2 dye; 5 MilliQ H2O 10 D9 + C1 Ligation (June 8/2010) 5 DNA; 2 dye; 5 MilliQ H2O Gel run for 80 minutes at 100V
Looks like everything was restricted. Ligations may work this time.
Follow-up: Ligation mixtures can be restriction tested and (if cutting works) transformed into DH5α cells.
Objective: Transform pDNA from distribution plates into DH5α cells to generate glycerol stocks for lab.
Method:
Remove DNA from the following locations on the 2010 distributions kits:- double terminator (B0010-B0010; AKA B0017) from kit plate 2 well 6K (pSB1A2)
- double terminator (B0010-B0012; AKA B0015) from kit plate 1 well 23L (pSB1AK3)
Transform into DH5α cells using transformation protocol, with pUC19 as positive control (1ng/µL), and water as negative control.
Results:
- Positive control: TMTC (too many to count) colonies
- B0010-B0010: 41 colonies
- B0010-B0012: 120 colonies
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
 "
"


 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]