Team:TU Munich/Project
From 2010.igem.org
(→Overall project) |
(→Overall project) |
||
| Line 59: | Line 59: | ||
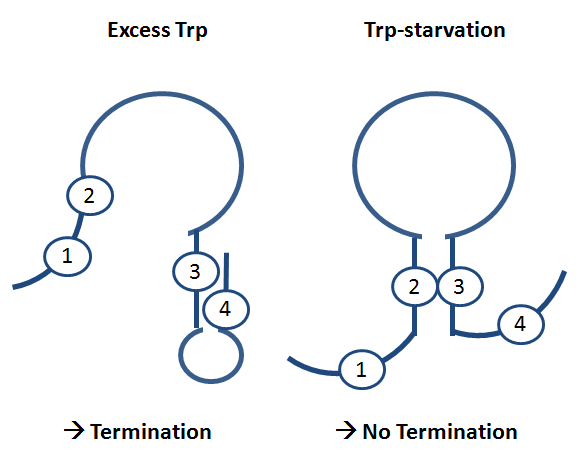
[[Image:TUM2010 attenuation.png|left|300 px|Principle of trp operon regulation in ''E. coli'']] | [[Image:TUM2010 attenuation.png|left|300 px|Principle of trp operon regulation in ''E. coli'']] | ||
| - | The concept on which we rely is a totally new on, based on the principle of [[ | + | The concept on which we rely is a totally new on, based on the principle of [[Glossary#Attenuation/ '''attenuation''']]. |
We applied this principle of RNA-RNA interaction based regulation to build switches based on antitermination. Note, that our systems works with termination on transcription level, so instead of stalling a ribosome, in our case the RNA-Polymerase falls off. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the '''signal'''), which binds to the transcription mRNA and anticipates the formation of a stem loop. So you basicly get a yes/no answer with a first '''switch''': If the small RNA piece is available, transcription will continue. If not, it will be terminated. It's that easy! <br> | We applied this principle of RNA-RNA interaction based regulation to build switches based on antitermination. Note, that our systems works with termination on transcription level, so instead of stalling a ribosome, in our case the RNA-Polymerase falls off. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the '''signal'''), which binds to the transcription mRNA and anticipates the formation of a stem loop. So you basicly get a yes/no answer with a first '''switch''': If the small RNA piece is available, transcription will continue. If not, it will be terminated. It's that easy! <br> | ||
Revision as of 19:21, 7 September 2010
|
||||||||||||||||
|
|
This is a template page. READ THESE INSTRUCTIONS.
You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples HERE.
You MUST have a team description page, a project abstract, a complete project description, a lab notebook, and a safety page. PLEASE keep all of your pages within your teams namespace.
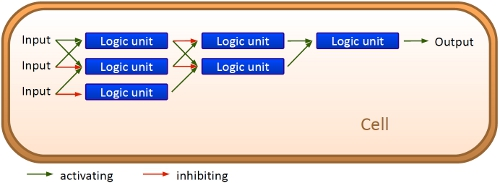
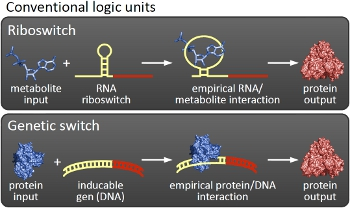
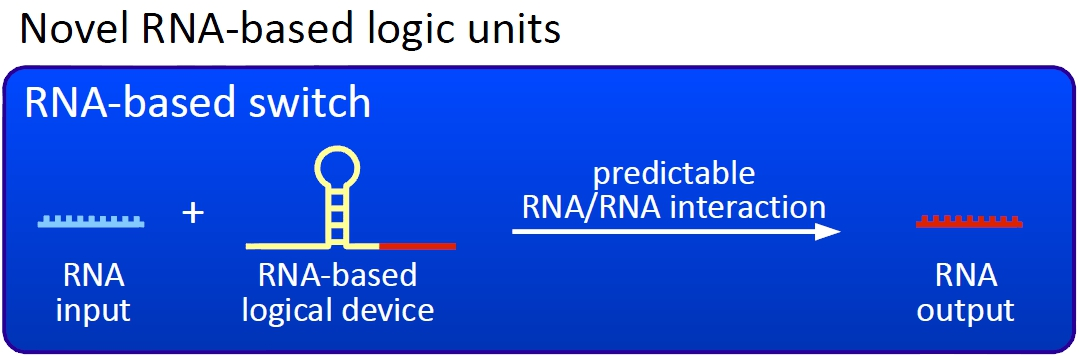
Overall projectWe, the TU Munich 2010 team, would like to change the usage and handling of Biobricks. Over the years so many teams spent time on evolving and constructing receptors and systems to detect a certain input that a variety of gorgeous oppurtunities is available so far. Nevertheless, up to now there are not real possibilities to link all those functionalities and built a network which can respond to many of those input signals in a highly differenciated way. We would like to provide a new system to control gene expression on RNA level which can be easily upscaled, which is capable of forming and/or/xor-links and offers for the first time the opportunity to built up complex and logical networks in E. coli cells. Isn't that great? The concept on which we rely is a totally new on, based on the principle of [[Glossary#Attenuation/ attenuation]]. We applied this principle of RNA-RNA interaction based regulation to build switches based on antitermination. Note, that our systems works with termination on transcription level, so instead of stalling a ribosome, in our case the RNA-Polymerase falls off. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the signal), which binds to the transcription mRNA and anticipates the formation of a stem loop. So you basicly get a yes/no answer with a first switch: If the small RNA piece is available, transcription will continue. If not, it will be terminated. It's that easy! Now, what's the great thing about our system? Well, we can construct a lot of those switches. The only things you need are complementary RNA sequences and one of them must form a stem loop big enough to terminate transcription (switch) if the other one (signal) is not there. Those RNA-RNA-interactions can easily be calculated and RNA structures can be predicted with high accuracy - in complete opposite to anything based on proteins. So we are easily capable of scaling up our switch and add a couple more of those guys into a bacteria cell (they are really small, both switch and signal RNA). Since we can use RNA switches to regulate RNA output, we can easily built up networks, with our RNA pieces as the major controlling element. In comparison to proteins, which are normally used for work like this, RNA is predictable in its structure, fast in production and fast in degeneration, so quick and time-related responses are possible, non-toxic in all cases, really cute, available in all shapes, consists only of a few sugars, phosphates, and four bases, there is an nearly endless pool of possible switches... You need more? Well, one more we would really like to mention: RNA can be generated using DNA. Big news? It was in the Watson/Crick era! Well, since RNA can be generated using DNA, you can really really easily code for our RNA switches using certain DNA sequences. DNA is stable and quite easy to get into cells and wait, Biobricks consists of DNA! So this is how we would like to revolutionize Biobricks. We developed switches based on a very easy, yet totally new principle, which can be just cloned in between known Biobricks and offer totally new possibilities in combining bricks and gene regulation! Check here for more information, a cute Java applet to form your own network and enjoy! Project DetailsPart 2The ExperimentsPart 3Results |
|||||||||||||||
 "
"