Team:Newcastle/12 July 2010
From 2010.igem.org
(→Materials and Protocol) |
(→Conditions) |
||
| Line 30: | Line 30: | ||
Please refer to [[Team:Newcastle/PCR|PCR]]. The melting temperature, Tm, of this experiment is 54°C. | Please refer to [[Team:Newcastle/PCR|PCR]]. The melting temperature, Tm, of this experiment is 54°C. | ||
| - | |||
| - | |||
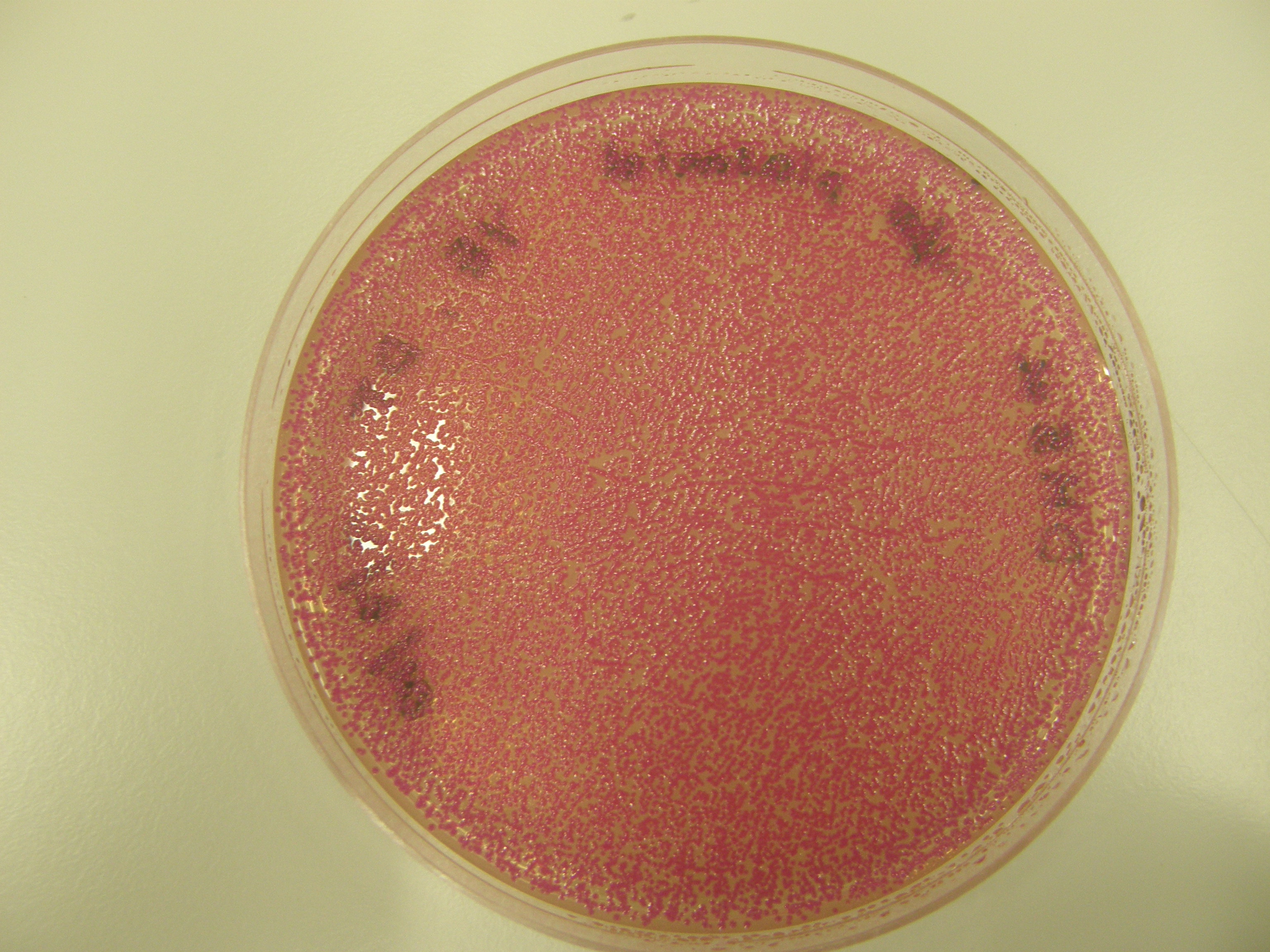
'''Note''' There are 2 possible reasons for red colonies formed: | '''Note''' There are 2 possible reasons for red colonies formed: | ||
Revision as of 15:34, 6 August 2010

| |||||||||||||
| |||||||||||||
Contents |
LacI BioBrick Construction
Aims
To use PCR to extract lacI (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP).
Summary:
- Colony PCR
- Streak plate
- Miniprep PCR - of colonies that worked
Materials
- PCR reagents
- Agar plates
- Gloves
- Wire loop
Colony PCR
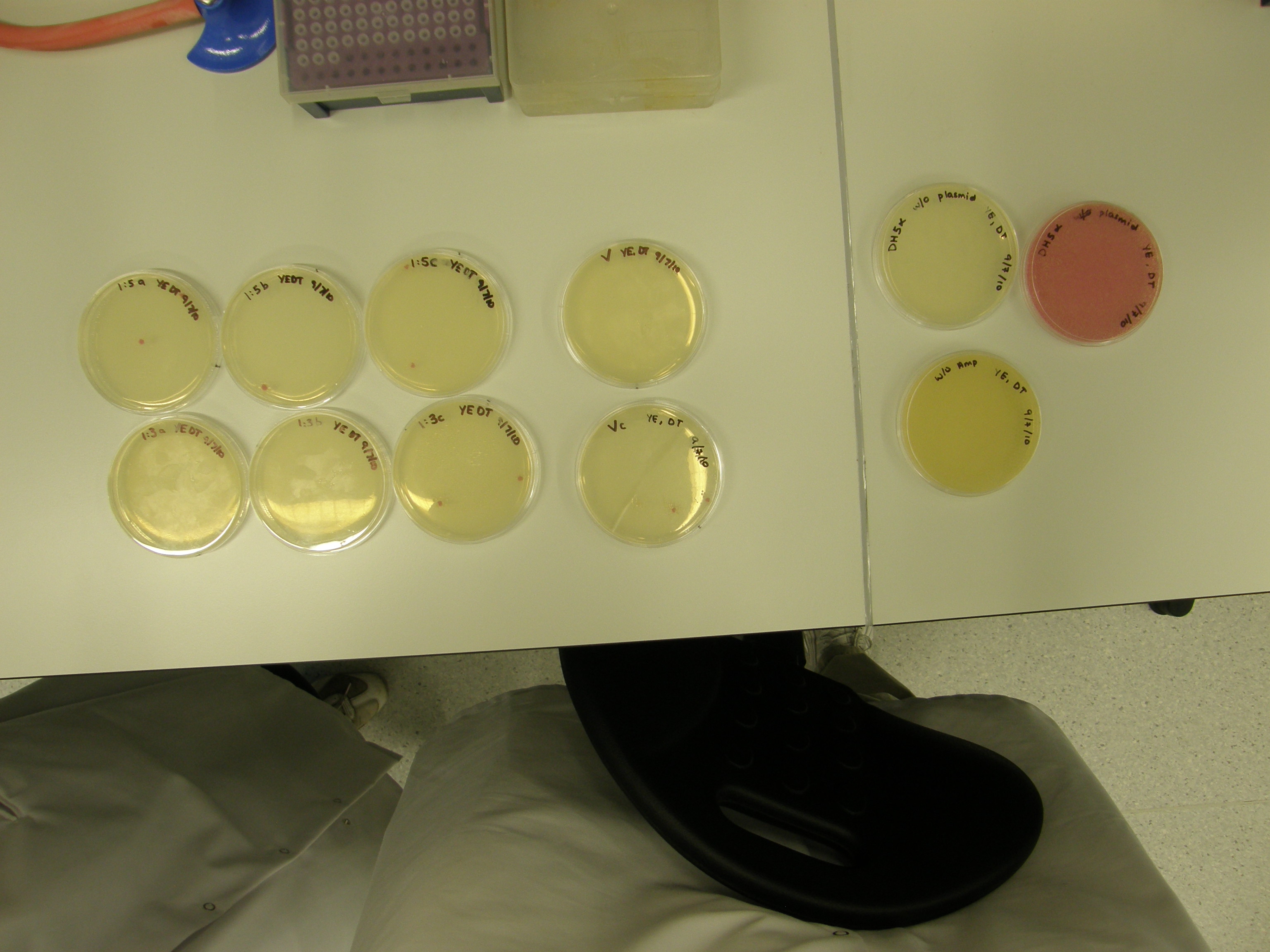
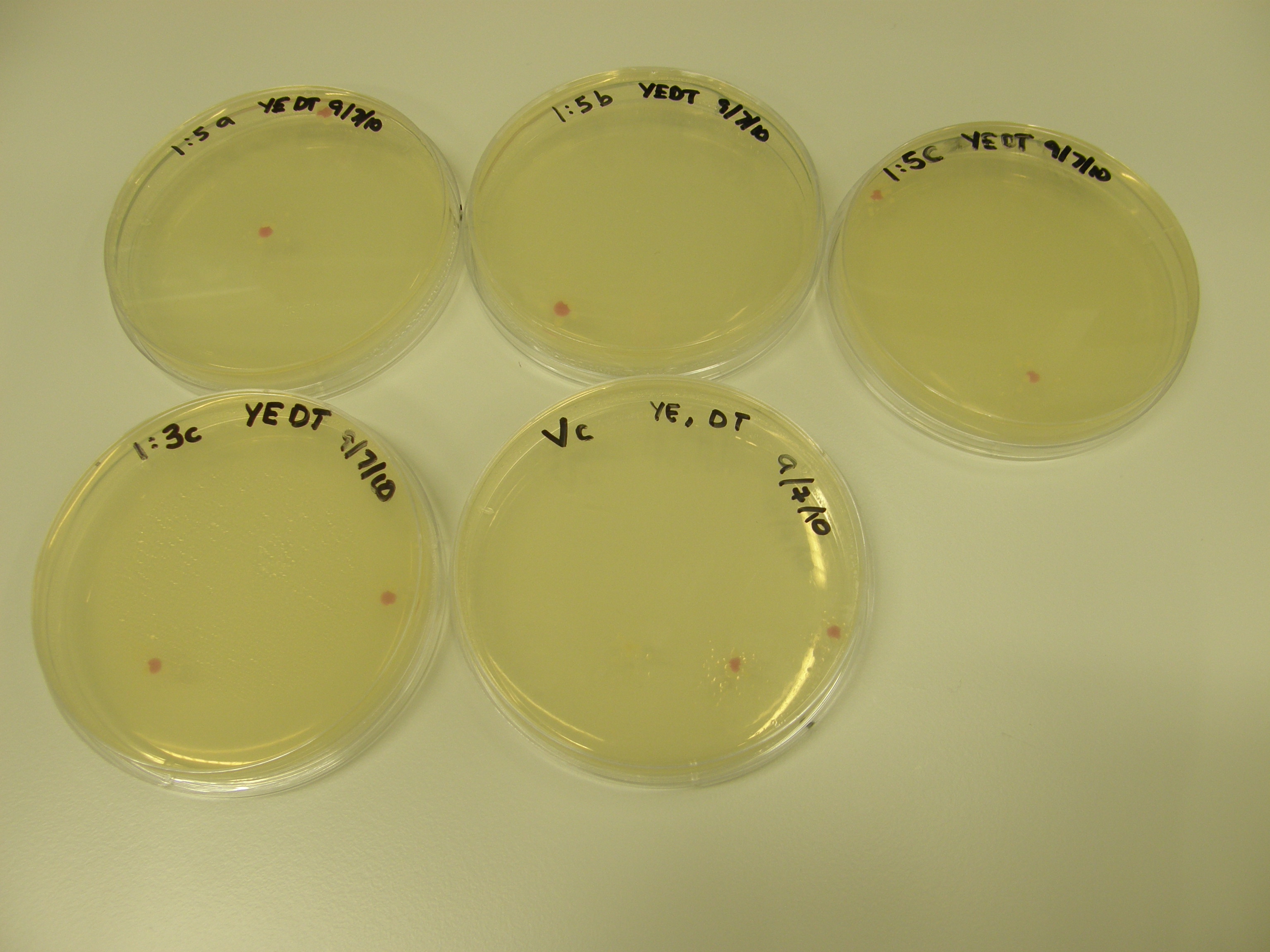
Of the 11 plates with colonies grown 7 were used for colony PCR and Streak plating. Colony PCR is less accurate than the mini prep however results can be seen quicker whereas the miniprep has a 1 day wait. The colonies selected selected were satellite colonies because ampicillin degraded???
7 Tubes were labelled 1-7 (+ a control)
The quantities for the PCR are as follows:
Materials and Protocol
Please refer to PCR. The melting temperature, Tm, of this experiment is 54°C.
Note There are 2 possible reasons for red colonies formed:
- partial digest - rejoins without insert
- with insert
We need to tell the difference between the two.
Note PCR is best when everything is kept on ice!
Note Melting temperatures of primers is important and can have a big effect on the product formed (Temperature used see Thursday)
Spread Plates
Overnight culture
The seven colonies that grew on the plates were cultured overnight. The protocol for growing an overnight culture was followed.
Tommorrow
We are going to take one of the colonies that have worked and possibly a whole plasmid prep.
 
|
 "
"