Team:Newcastle/End of crack & signalling system
From 2010.igem.org
Swoodhouse (Talk | contribs) (→End of crack cell-signalling system) |
Swoodhouse (Talk | contribs) (→Subtilin immunity BioBrick) |
||
| Line 12: | Line 12: | ||
==Subtilin immunity BioBrick== | ==Subtilin immunity BioBrick== | ||
| - | The spaIFEG gene cluster encodes two separate subtilin immunity systems, both present in ATCC 6633. Together they provide a high degree of subtilin resistance. | + | The spaIFEG gene cluster encodes two separate subtilin immunity systems, both present in ATCC 6633. Together they provide a high degree of subtilin resistance. |
| + | |||
| + | [[Media:Newcastle CloningStrategy_BioBrick_Subtilin_Immunity.pdf|Cloning strategy]] | ||
==Subtilin production BioBrick== | ==Subtilin production BioBrick== | ||
Revision as of 13:27, 10 August 2010

| |||||||||||||
| |||||||||||||
Contents |
End of crack cell-signalling system
We aim to use a subtilin-based cell signalling system to trigger calcium carbonate precipitation and filament formation once our bacteria have reached a sufficient density inside a microcrack.
Subtilin is a lantibiotic peptide produced naturally by Bacillus subtilis ATCC 6633 but not by Bacillus subtilis 168. We aim to produce two BioBricks: one for production of subtilin, and, since subtilin is a lantibiotic, one for subtilin immunity. A subtilin-sensing BioBrick was produced by Newcastle's 2008 iGEM team.
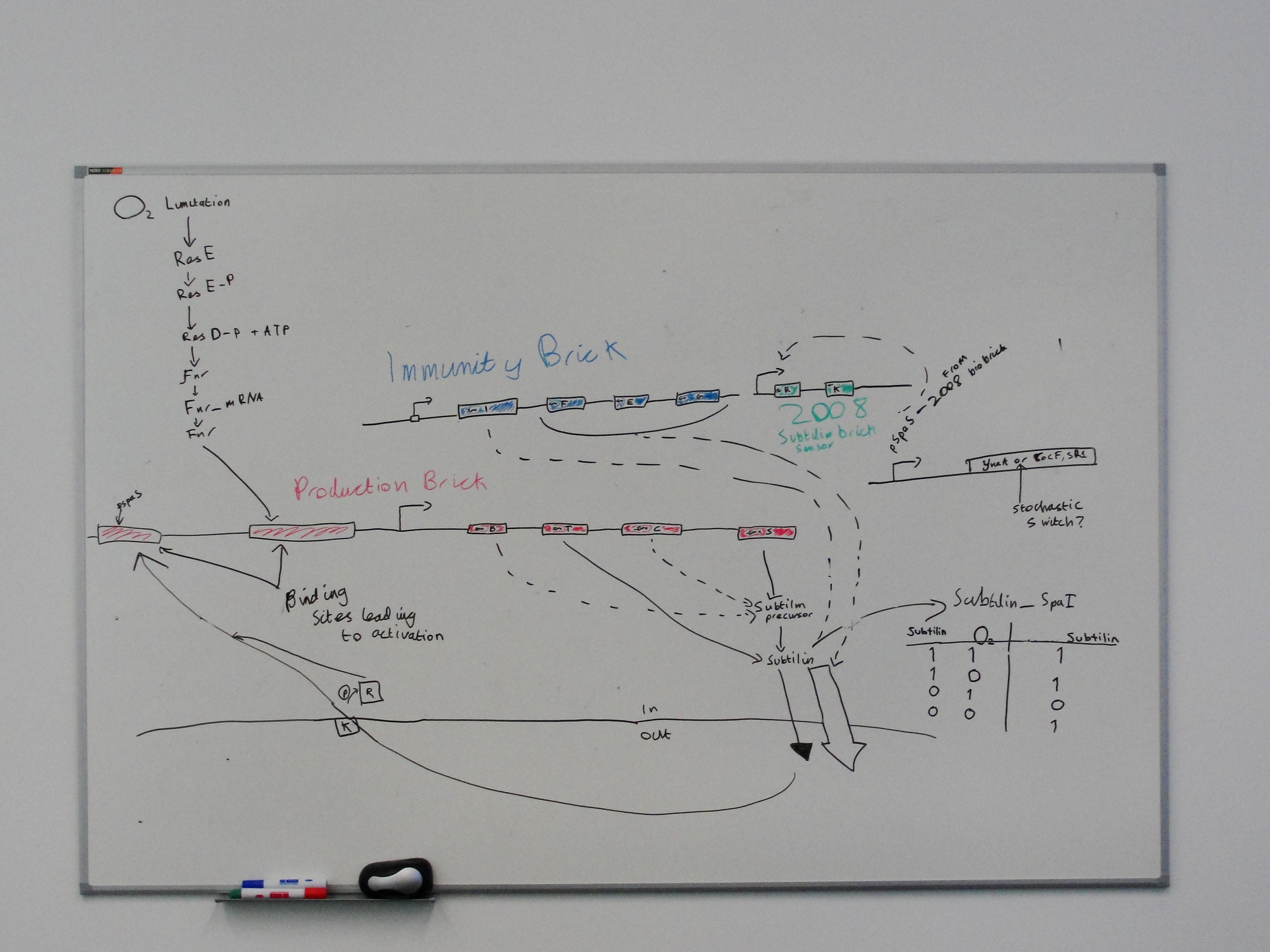
Subtilin immunity BioBrick
The spaIFEG gene cluster encodes two separate subtilin immunity systems, both present in ATCC 6633. Together they provide a high degree of subtilin resistance.
Subtilin production BioBrick
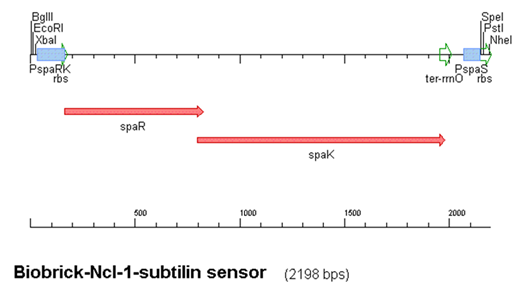
2008 Subtilin-sensing BioBrick
- A type 1 antimicrobial peptide [AMP] or lantibiotic produced by B. subtilis
- NB. Lantibiotic = peptide-derived antibiotics with high antimicrobial activity against various Gram-positive bacteria, including pathogenic bacteria such as propionibacteria, staphylococci, clostridia, enterococci and streptococci.
- NB. Subtilin not same as Subtilisin!!!!!
- Production of subtilin involves the spa gene cluster encompassing 10 genes, spaBTCSIFEGRK
The construct above contains:
- spaRK promotor
- spaR (subtilin peptide antibiotic Regulation) - the 220 amino acid product of this gene usually regulates the downstream production of subtilin antibiotic. It has an N-terminal domain that can be phosphorylated and a C-terminal domian that has DNA binding properties [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T0M-4D4XNMM-4&_user=224739&_coverDate=09%2F01%2F2004&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000014659&_version=1&_urlVersion=0&_userid=224739&md5=1f421f180a48e2d68b86da579cc7f920|1]
- spaK (subtilin peptide antibiotic Kinase) - this gene codes for a 325 amino acid histadine kinase peptide that phosphorylates the N-terminus of SpaR [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T0M-4D5KTFV-1&_user=224739&_coverDate=09%2F01%2F2004&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000014659&_version=1&_urlVersion=0&_userid=224739&md5=8d450f455463638579798872811ae5c0|2]. This activates the DNA binding ability of the C-terminus of SpaR, which in turn initiates transcription of the downstream gene. In the case of our construct, this gene is gfp.
- rrnO transcriptional terminator
- spaS promotor - a strong promotor inducible by upstream activation of spaRK. It can be placed in front any gene to regulate its activity.
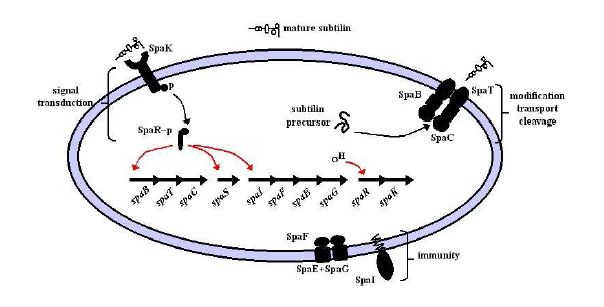
Production and Immunity Bricks
- spaBTCS genes are required for production.
- spaIFEG genes are required for immunity.
- spaB and C are modifiers of the subtilin precursor.
- spaT is a transporter.
- spaS is the gene coding for the subtilin precursor peptide.
- spaI 's gene product sequesters subtilin.
- spaFEG genes code for transporters.
Image from Modeling Subtilin production in Bacillus subtilis Using Stochastic Hybrid Systems. Hu et al.
Biochemical pathway
 
|
 "
"