Team:TU Delft/26 July 2010 content
From 2010.igem.org
(→Alkane Degradation) |
|||
| Line 130: | Line 130: | ||
|1 | |1 | ||
|rubA3 | |rubA3 | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |PvuI | + | |0.5μl PvuI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 138: | Line 138: | ||
|2 | |2 | ||
|rubA4 | |rubA4 | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |PvuI | + | |0.5μl PvuI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 146: | Line 146: | ||
|3 | |3 | ||
|rubR | |rubR | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |EcoRI | + | |0.5μl EcoRI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 154: | Line 154: | ||
|4 | |4 | ||
|ladA | |ladA | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |PvuI | + | |0.5μl PvuI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 162: | Line 162: | ||
|5 | |5 | ||
|ADH | |ADH | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |EcoRI | + | |0.5μl EcoRI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 170: | Line 170: | ||
|6 | |6 | ||
|ALDH | |ALDH | ||
| - | |XbaI | + | |1μl XbaI |
| - | |PstI | + | |1μl PstI |
| - | |PvuI | + | |0.5μl PvuI |
|3 (Biolabs) | |3 (Biolabs) | ||
|✓ | |✓ | ||
| Line 178: | Line 178: | ||
|7 | |7 | ||
|J61100 | |J61100 | ||
| - | |SpeI | + | |1μl SpeI |
| - | |PstI | + | |1μl PstI |
| - | | - | ||
|2 (Biolabs) | |2 (Biolabs) | ||
| Line 186: | Line 186: | ||
|8 | |8 | ||
|J61101 | |J61101 | ||
| - | |SpeI | + | |1μl SpeI |
| - | |PstI | + | |1μl PstI |
| - | | - | ||
|2 (Biolabs) | |2 (Biolabs) | ||
| Line 194: | Line 194: | ||
|7 | |7 | ||
|J61107 | |J61107 | ||
| - | |SpeI | + | |1μl SpeI |
| - | |PstI | + | |1μl PstI |
| - | | - | ||
|2 (Biolabs) | |2 (Biolabs) | ||
|✓ | |✓ | ||
Revision as of 14:13, 27 July 2010
Emulsification Assay
We have been looking for a robust protocol for the characterization of the emulsifiers for quite some time now. Somehow most literature refers to spectrophotometric assays based on turbidity after mixing. This a somewhat arbitrary method, and so far we were unable to get reproducible results. Today we decided to explore this method once more, using a lot of different conditions.
The method is based on a simple article in Biochemical and Biophysical Methods: Spectrophotometric method for quantitative determination of nonionic, ionic and zwitterionic detergents by Rajakumari et al. 2006.
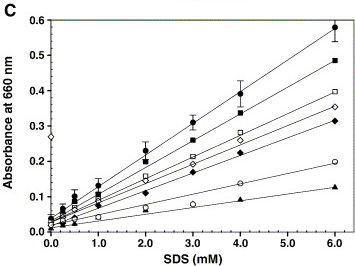
The article shows the graph on the right, which we tried to reproduce. However, instead of triolein we used hexane. And the buffer used was 200 mM Tris-HCl, pH 8.
| # | [SDS] (mM) | Absorbance 660 nm (AU) |
| 1 | 10 | 0.598 |
| 2 | 8 | 0.425 |
| 3 | 6 | 0.830 |
| 4 | 4 | 0.432 |
| 5 | 2 | 0.379 |
| 6 | 0 | 0.001 |
This looked promising, so the experiment was carried out once more in triplo and with proper controls.
| # | [SDS] (mM) | Serie 1 | Serie 2 | Serie 3 | W/O Hexane |
| 1 | 8 | 0.640 | 0.439 | 0.622 | 0.011 |
| 2 | 6 | 0.454 | 0.389 | 0.410 | 0.014 |
| 3 | 5 | 0.514 | 0.539 | 0.440 | 0.023 |
| 4 | 4 | 0.365 | 0.415 | 0.449 | 0.022 |
| 5 | 3 | 0.346 | 0.485 | 0.478 | 0.028 |
| 6 | 2 | 0.413 | 0.404 | Sorry, dropped the sample :( | 0.024 |
| 7 | 1 | 0.399 | 0.500 | 0.388 | 0.011 |
| 8 | 0 | 0.003 | 0.007 | 0.004 | 0.002 |
As you can see in the Table above, the measurements are kind of random. The way in which you pipet also influences the measurement a lot. That is why we decided to vortex the fluid in the cuvet, and take lower concentrations.
This experiment was also done in triplo:
The conclusion is that hexane dissolves much easier than triolein. This curve might be useful in measuring the emulsification capacity of our proteins. So, next will try to measure the emulsification capacity of our positive control strain P. putida.
Alkane Degradation
Today we're going to try to make the BioBricks again, but using a different method. By cutting the RBSs with SpeI and PstI, so not removing the RBS from the plasmid, and the other piece (gene) with XbaI and PstI, this piece can be inserted into the other plasmid using a 2-way ligation. To prevent the 'insert' plasmid from reclosing and extra cut was done in the plasmid.
| # | Fragment | Enzyme 1 | Enzyme 2 | Enzyme 3 (extra cut) | Used Buffer | BSA |
| 1 | rubA3 | 1μl XbaI | 1μl PstI | 0.5μl PvuI | 3 (Biolabs) | ✓ |
| 2 | rubA4 | 1μl XbaI | 1μl PstI | 0.5μl PvuI | 3 (Biolabs) | ✓ |
| 3 | rubR | 1μl XbaI | 1μl PstI | 0.5μl EcoRI | 3 (Biolabs) | ✓ |
| 4 | ladA | 1μl XbaI | 1μl PstI | 0.5μl PvuI | 3 (Biolabs) | ✓ |
| 5 | ADH | 1μl XbaI | 1μl PstI | 0.5μl EcoRI | 3 (Biolabs) | ✓ |
| 6 | ALDH | 1μl XbaI | 1μl PstI | 0.5μl PvuI | 3 (Biolabs) | ✓ |
| 7 | J61100 | 1μl SpeI | 1μl PstI | - | 2 (Biolabs) | ✓ |
| 8 | J61101 | 1μl SpeI | 1μl PstI | - | 2 (Biolabs) | ✓ |
| 7 | J61107 | 1μl SpeI | 1μl PstI | - | 2 (Biolabs) | ✓ |
 "
"