Team:Calgary/20 July 2010
From 2010.igem.org
H.dastidar (Talk | contribs) |
Raidakhwaja (Talk | contribs) |
||
| Line 9: | Line 9: | ||
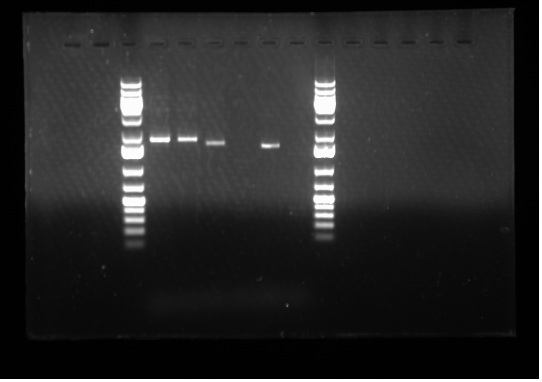
Today I set up and ran two 1% gels: one for my PCR products from yesterday and the second one for Emily's PCR products- both of which were done to test whether our primers are working or not. Please see the gel to the side: | Today I set up and ran two 1% gels: one for my PCR products from yesterday and the second one for Emily's PCR products- both of which were done to test whether our primers are working or not. Please see the gel to the side: | ||
As we can see in the gel electrophoresis image, the expected results has been obtained. For example, Lane 8 ladder contains only the Lux0047E gene where as Lane 6 has J23002 + Lux0047E, hence the band in Lane 8 goes slightly further relative to Lane 6 because it is a shorter band. Lanes 4 and 5 show exactly the same band because they are the same genes in the same plasmid backbone. Furthermore, Lane 9 shows no band because it is the Master Mix and our negative control. Therefore, it can be concluded that the '''primers are functional.''' | As we can see in the gel electrophoresis image, the expected results has been obtained. For example, Lane 8 ladder contains only the Lux0047E gene where as Lane 6 has J23002 + Lux0047E, hence the band in Lane 8 goes slightly further relative to Lane 6 because it is a shorter band. Lanes 4 and 5 show exactly the same band because they are the same genes in the same plasmid backbone. Furthermore, Lane 9 shows no band because it is the Master Mix and our negative control. Therefore, it can be concluded that the '''primers are functional.''' | ||
| - | + | ||
| + | Apart from the Wet Lab, I've also helped Jeremey with the summary of our Lethbridge Workshop. | ||
<u> Jeremy </u> | <u> Jeremy </u> | ||
Revision as of 22:36, 20 July 2010

Tuesday July 20, 2010
Raida
Today I set up and ran two 1% gels: one for my PCR products from yesterday and the second one for Emily's PCR products- both of which were done to test whether our primers are working or not. Please see the gel to the side: As we can see in the gel electrophoresis image, the expected results has been obtained. For example, Lane 8 ladder contains only the Lux0047E gene where as Lane 6 has J23002 + Lux0047E, hence the band in Lane 8 goes slightly further relative to Lane 6 because it is a shorter band. Lanes 4 and 5 show exactly the same band because they are the same genes in the same plasmid backbone. Furthermore, Lane 9 shows no band because it is the Master Mix and our negative control. Therefore, it can be concluded that the primers are functional.
Apart from the Wet Lab, I've also helped Jeremey with the summary of our Lethbridge Workshop.
Jeremy
Today I completed the ligation and transformation of the I0500+E0034 plasmid switch. I also ran the PCR products and the results came back negative. So I set up another PCR and Gel.
Patrick
Today I helped plasmid prep the degP promoter and cpxR promoter on a GFP and RFP reporter device each. I also continued looking to literature regarding high and low-copy plasmids for the modelling project.
Himika
Today I plated the construction that I did last night. I also overnight cultured the construction from last night by inoculating from the cells to be plated. Tomorrow I will plasmid prep the cultures. I also looked into the literature regarding the plasmid which will be presented to Paul on Thursday.
No notebook page exists for this date. Sorry! "
"