Team:SDU-Denmark/labnotes2
From 2010.igem.org
m (→Transformation of K081005 in pSB1A2 (constitutive promoter and RBS combined),R0011 in pSB1A2, pSB3C5 w. J04450 and pSB3T5 w. J04450 in Top 10 E.Coli) |
m (→Ligation of flhD/C and pSB1C3) |
||
| Line 261: | Line 261: | ||
</tr> | </tr> | ||
</table> <br><br> | </table> <br><br> | ||
| - | Ligation mixtures were not run on gel but were directly used for transformation<br> | + | Ligation mixtures were not run on gel but were directly used for [https://2010.igem.org/Team:SDU-Denmark/labnotes2#Transformation_of_flhD.2FC_in_pSB1C3_and_test_plasmid_in_Top_10_E.Coli transformation]<br> |
| + | surplus ligation mixture was stored at -20 degrees as L1, L2 and L3.<br> | ||
--[[User:Tipi|Tipi]] 18:15, 19 July 2010 (UTC) | --[[User:Tipi|Tipi]] 18:15, 19 July 2010 (UTC) | ||
Revision as of 16:27, 20 July 2010
Lab notes (7/19 - 7/25)
Contents |
Group: Flagella
Annealing of the two mutated strands of FlhDC
Experiment done by: Sheila
Date: July 19th
Protocol: CP1.1
Method: PCR of the two mutated strands of the FlhDC operon
Notes: To samples were run at two different temperatures: 56,1˚C and 64,5˚C respectively.
Polymerase used: Pfu
Primers used: None, as the two strands are supposed to anneal to each other
Results:
Gel extraction of flhD/C
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.1
Experiment done by:Maria
Notes:
70uL of flhD/C PCR product from Amplification of flhD/C was loaded onto a 1.5 agarose gel and extracted according to protocol.
DNA was eluted in 20uL elution buffer.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhDC 1 |
19.82 |
2.43 |
0.08 |
| flhDC 2 |
25.24 |
2.22 |
0.12 |
Analysis:
Nanodrop measurements indicated a possible contamination. However the DNA was pooled and used for Digestion
--Tipi 17:22, 19 July 2010 (UTC)
Digestion of flhD/C and pSB1A2 with EcoRI SpeI
Start date: 19/07 End date: 19/07
Methods: Digestion, Gel electrophoresis
Protocol:RD1.1
Experiment done by:Maria
Notes:
purified pSB1C3 (tube 18 blue) and flhD/C from Gel extraction was digested with EcoRI and SpeI.
Restriction mixture:
| flhD/C |
pSB1C3 |
|
| H2O |
30uL |
24uL |
| EcoRI |
4uL |
2uL |
| SpeI |
4uL |
2uL |
| FD green buffer |
8uL |
4uL |
| DNA |
38uL |
10uL |
| total vol. |
84 |
42 |
samples were loaded onto a 1.5 agarose gel. Gene ruler red were used as marker.
Results:
Gel electrophoresis:
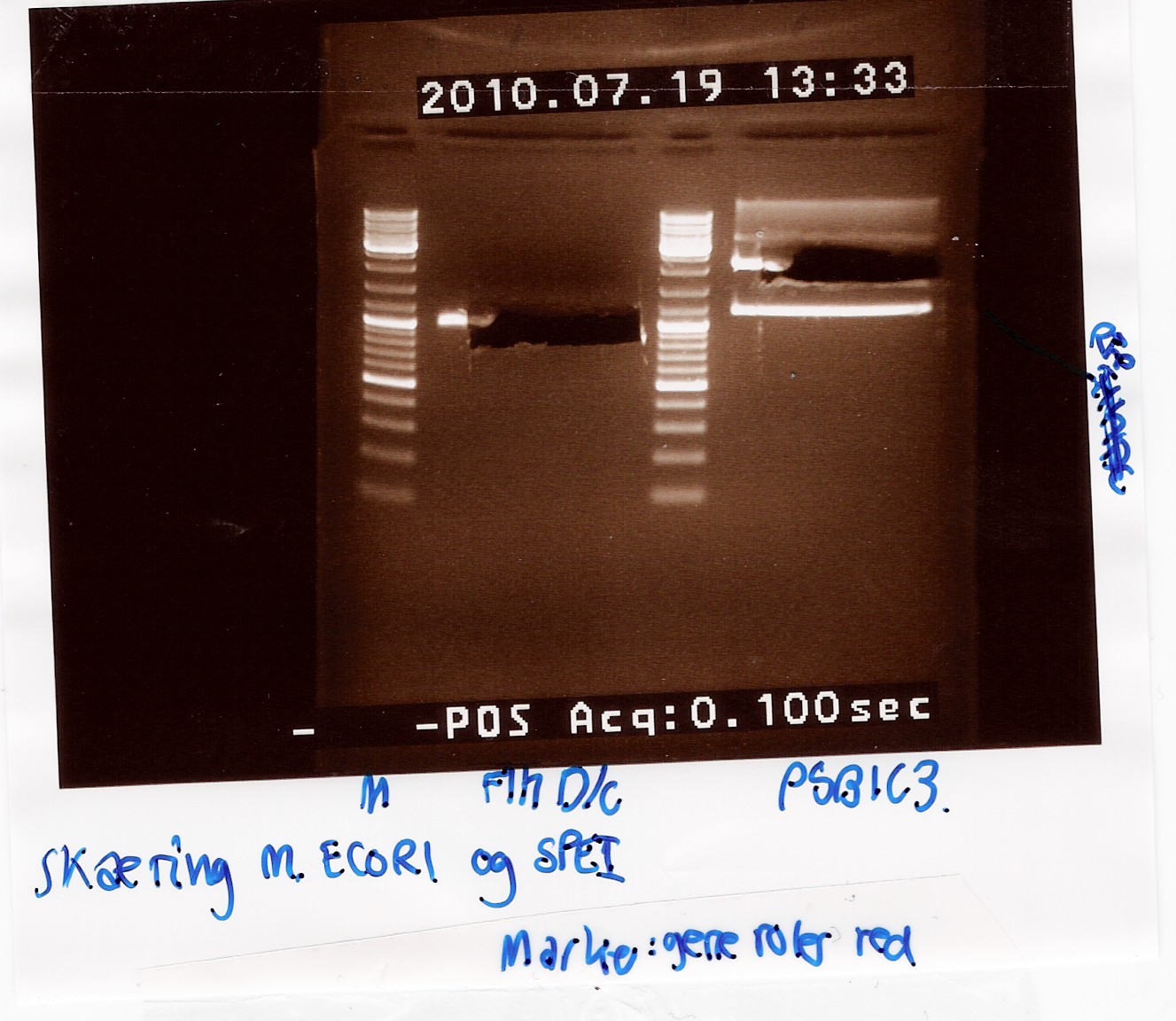
Analysis:
In lane 2 containing pSB1C3 2 bands are detected indicating a succesful digestion of the plasmid (the band at 1000 bp corresponds to J04450). A succesful digestion of the flhD/C cannot be concluded from the gel. However both bands was excised and extracted from gel (Gel extraction)
--Tipi 17:42, 19 July 2010 (UTC)
Gel extraction of digested flhD/C and pSB1C3
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.2
Experiment done by:Maria
Notes:
Digested flhD/C and pSB1C3 from Digestion were extracted from gel according to protocol.
DNA was eluted in 20uL H20.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhD/C |
18.18 |
4.59 |
0.18 |
| pSB1C3 |
14.30 |
1.89 |
0.04 |
Analysis:
nanodrop measurements indicated contamination. However both samples were used for Ligation.
--Tipi 17:57, 19 July 2010 (UTC)
Ligation of flhD/C and pSB1C3
Start date: 19/07 End date: 20/07
Methods: Ligation
Protocol:LG1.2
Experiment done by:Maria
Notes:
3 ligation reactions was prepared.
| LG1 |
LG2 |
LG3 |
|
| 10x T4 ligase buffer |
2uL |
2uL |
2uL |
| flhD/C |
5uL |
5uL |
5uL |
| pSB1C3 |
1uL |
2.8uL |
5uL |
| H20 |
11uL |
9.2uL |
7uL |
| T4 ligase |
1uL |
1uL |
1uL |
Ligation mixtures were not run on gel but were directly used for transformation
surplus ligation mixture was stored at -20 degrees as L1, L2 and L3.
--Tipi 18:15, 19 July 2010 (UTC)
Group: Photosensor
Group: Retinal
Transformation of K081005 in pSB1A2 (constitutive promoter and RBS combined),R0011 in pSB1A2, pSB3C5 w. J04450 and pSB3T5 w. J04450 in Top 10 E.Coli
Start date: 19/07 End date: 20/07
Methods: ON culture, making competent cells, transformation
Protocol:CC1.1 TR1.1
Experiment done by: Maria
Notes:ON colony was made of 110 ml lb medium inoculated with a top10 coloni.
| Time |
Optical density |
| 8:12 |
2.9 |
| 8:17 |
0.02 |
| 9:17 |
0.035 |
| 10:17 |
0.204 |
| 10:40 |
0.380 |
| 10:50 |
0.49 |
pSB1A2 w. R0011 and pSB1A2 w. K081005 was plated with 150uL on plates containing LA, LA + Amp, LA + Tetracycline, LA + Chloramphenicol and LA + Kanamycine.Upconcentration of these samples was not made. pSB3T5 w. J04450 and pSB3C5 w. J04450 was plated according to protocol
Results:
Analysis:
Both pSB3T5 and pSB3C5 was succesfully transformed, and ON cultures with appropiate antibiotics were made for mini-prep.
For pSB1A2 w. R0011 only 6 colonies was observed on the LA+amp plate.
For pSB1A2 w. K081005 only 4 colonies was observed on the LA+amp plate.
All 10 colonies were used for Coloni PCR.
--Tipi 16:33, 19 July 2010 (UTC)
Transformation of flhD/C in pSB1C3 and test plasmid in Top 10 E.Coli
Start date: 20/07 End date: 19/07
Methods: ON culture, making competent cells, transformation
Protocol:CC1.1 TR1.1
Experiment done by: Maria
Notes:
Ligated flhD/C from Ligation and test plasmid from Whatman was transformed.
ON colony was made of 25 ml lb medium inoculated with a top10 coloni.
| Time |
Optical density |
| 8:17 |
3.5 |
| 8:20 |
0.02 |
| 9:15 |
0.035 |
| 10:30 |
0.222 |
| 10:52 |
0.350 |
| 11:05 |
0.52 |
Compotent cells were washed using 10mL 50mM CaCl2.
3 parallel transformations were carried out for L1, L2 and L3 respectively (see Ligation). L1.1, L1.2, L2.1, L2.2 and L3.1 were transformed using compotent cells from 19/7 (Compotent cells).
Prior to transformation test plasmid was washed with 10xTE pH 8.0.
--Tipi 15:19, 20 July 2010 (UTC)
Coloni PCR of R0011 in pSB1A2 and K081005 in pSB1A2
Start date: 20/07 End date: 20/07
Methods: Coloni PCR and gel electrophoresis
Protocol:CP1.2
Experiment done by: Maria and LC
Notes:
6 colonies transformed with pSB1A2 w. R0011 and 4 colonies transformed with pSB1A2 w. K081005 (see Transformation) was used for coloni PCR.
Colonies with K081005 was denoted A1, A2 A3 and A4 respectively.
Colonies with R0011 was denotes B1, B2, B3, B4, B5 and B6 respectively
PCR was made with only half the amount of premix.
Premix for 12 PCR reactions:
| 10xtaq buffer |
30uL |
| MgCl2 |
12uL |
| VF2 |
12uL |
| VR |
12uL |
| dNTP |
6uL |
| H2O |
42uL |
| Total vol. |
114uL |
0.5uL Taq polymerase from ampliqon was used.
Taq PCR program:
| 1:Start |
94C |
2 min |
| 2: Denaturing |
94C |
1 min |
| 3: Annealing |
55C |
1 min |
| 4: Elongation |
72C |
30 s |
| 5: |
GO TO |
rep. 29x |
| 6: End |
72C |
3 min |
| 7: Hold |
4C |
PCR product was loaded onto a 2% agarose gel. Gene ruler 100bp DNA ladder was used as marker.
Results:
Analysis:
--Tipi 16:21, 20 July 2010 (UTC)
Include column content here.
 "
"