Team:Imperial College London/Lab Diaries/Vectors team
From 2010.igem.org
(table formatting) |
m (table formatting) |
||
| Line 144: | Line 144: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis and extraction of 5' ins</li> | <li>Gel analysis and extraction of 5' ins</li> | ||
| Line 157: | Line 157: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Replica plating and colony PCR of transformed colonies (containing 5' dif in pSB1C3)</li> | <li>Replica plating and colony PCR of transformed colonies (containing 5' dif in pSB1C3)</li> | ||
| Line 165: | Line 165: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation of overnight ligations: 5' ins, dif with pSB1C3 and pveg, SpecR-T and pSB1C3 </li> | <li>Transformation of overnight ligations: 5' ins, dif with pSB1C3 and pveg, SpecR-T and pSB1C3 </li> | ||
| Line 172: | Line 172: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Replica plating and colony PCR of: 5' ins, dif with pSB1C3 and pveg, SpecR-T and pSB1C3 </li> | <li>Replica plating and colony PCR of: 5' ins, dif with pSB1C3 and pveg, SpecR-T and pSB1C3 </li> | ||
| Line 181: | Line 181: | ||
<tr> | <tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>PCR purification of PSB1C3 vector </li> | <li>PCR purification of PSB1C3 vector </li> | ||
| Line 190: | Line 190: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Ligation of 5' ins and dif with pSB1C3 - a bench ligation (1 hour) and an overnight ligation were set up </li> | <li>Ligation of 5' ins and dif with pSB1C3 - a bench ligation (1 hour) and an overnight ligation were set up </li> | ||
| Line 199: | Line 199: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Overnight ligation of pveg and SpecR-T with pSB1C3 </li> | <li>Overnight ligation of pveg and SpecR-T with pSB1C3 </li> | ||
| Line 205: | Line 205: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of colony PCR products from the transformations</li> | <li>Gel analysis of colony PCR products from the transformations</li> | ||
| Line 300: | Line 300: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up overnight ligations for standard assembly (BBA) and 3A cloning (3A) of dif P</li> | <li>Set up overnight ligations for standard assembly (BBA) and 3A cloning (3A) of dif P</li> | ||
| Line 309: | Line 309: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Restriction digestion of 3' ins in the AK3 vector ( Using Xba and Pst) for 3A | <li>Restriction digestion of 3' ins in the AK3 vector ( Using Xba and Pst) for 3A | ||
| Line 316: | Line 316: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<ul> | <ul> | ||
| Line 324: | Line 324: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Replica plating of transformed colonies having dif P with 3' ins in AK3 -45 sigle colonies were plated</li> | <li>Replica plating of transformed colonies having dif P with 3' ins in AK3 -45 sigle colonies were plated</li> | ||
| Line 331: | Line 331: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of colony PCR from yesterday (first 15 replica plated colonies</li> | <li>Gel analysis of colony PCR from yesterday (first 15 replica plated colonies</li> | ||
| Line 338: | Line 338: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Miniprep of 4 overnight cultures - diff P with 3' ins in AK3; colonies 4,5,7 & 9</li> | <li>Miniprep of 4 overnight cultures - diff P with 3' ins in AK3; colonies 4,5,7 & 9</li> | ||
| Line 346: | Line 346: | ||
<tr> | <tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of PCR purified 3' ins in AK3 and gel purified 3' diff P to work out ratios for the liagation </li> | <li>Gel analysis of PCR purified 3' ins in AK3 and gel purified 3' diff P to work out ratios for the liagation </li> | ||
| Line 359: | Line 359: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation of E.Coli with ligates from yesterday in AmpR;dif P with 3' ins in AK3 (insert and vector) and AK3 containing 3' ins vector only</li> | <li>Transformation of E.Coli with ligates from yesterday in AmpR;dif P with 3' ins in AK3 (insert and vector) and AK3 containing 3' ins vector only</li> | ||
| Line 369: | Line 369: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of gel extracted dif P and 3' ins (inserts) and pSB1C3 (vector) to work out ratios for the ligation </li> | <li>Gel analysis of gel extracted dif P and 3' ins (inserts) and pSB1C3 (vector) to work out ratios for the ligation </li> | ||
| Line 379: | Line 379: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up overnight 5 ml cultures containing dif P with 3' ins in AK3 for miniprep tomorrow - 4 cultures were set up by looking at the gel this morning; 2 positive looking (4 & 7), 1 negative (5) and one containing nothing (9</li> | <li>Set up overnight 5 ml cultures containing dif P with 3' ins in AK3 for miniprep tomorrow - 4 cultures were set up by looking at the gel this morning; 2 positive looking (4 & 7), 1 negative (5) and one containing nothing (9</li> | ||
| Line 385: | Line 385: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Diagnostic digests of minipreps containing dif P with 3' ins in AK3 - Two digests : One with Spe & Pst and other with Xba & Spe </li> | <li>Diagnostic digests of minipreps containing dif P with 3' ins in AK3 - Two digests : One with Spe & Pst and other with Xba & Spe </li> | ||
| Line 422: | Line 422: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of Mini preps and diagnostic digests from Saturday - Colony 4 looked best</li> | <li>Gel analysis of Mini preps and diagnostic digests from Saturday - Colony 4 looked best</li> | ||
| Line 428: | Line 428: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Midiprep of Colony 4 - concentration 110 ng/ul </li> | <li>Midiprep of Colony 4 - concentration 110 ng/ul </li> | ||
| Line 434: | Line 434: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<ul> | <ul> | ||
| Line 442: | Line 442: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation with overnight ligations </li> | <li>Transformation with overnight ligations </li> | ||
| Line 448: | Line 448: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>The transformations were highly successful!! The Vector only plate showed no colonies and the Insert & Vector plate showed many individual colonies </li> | <li>The transformations were highly successful!! The Vector only plate showed no colonies and the Insert & Vector plate showed many individual colonies </li> | ||
| Line 457: | Line 457: | ||
</tr> | </tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up 200 ml overnight culture of colony 4 (containing Dif P and K09) for midiprep tomorrow </li> | <li>Set up 200 ml overnight culture of colony 4 (containing Dif P and K09) for midiprep tomorrow </li> | ||
| Line 466: | Line 466: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Digests set up for Vector (SpecR) with Spe & Pst and Insert (dif P & K09)with Xba & Ps</li> | <li>Digests set up for Vector (SpecR) with Spe & Pst and Insert (dif P & K09)with Xba & Ps</li> | ||
| Line 475: | Line 475: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Dephosphorylation of vector </li> | <li>Dephosphorylation of vector </li> | ||
| Line 482: | Line 482: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Both ligations (Insert & Vector and Vector only) were plated in CmR and incubated overnight </li> | <li>Both ligations (Insert & Vector and Vector only) were plated in CmR and incubated overnight </li> | ||
| Line 488: | Line 488: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of colony PCR products </li> | <li>Gel analysis of colony PCR products </li> | ||
| Line 524: | Line 524: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Colony PCR of 10 transformed C-Spec colonies</li> | <li>Colony PCR of 10 transformed C-Spec colonies</li> | ||
| Line 531: | Line 531: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Miniprep of C-Spec colonies 2, 5 & 9 in SpecR and CmR</li> | <li>Miniprep of C-Spec colonies 2, 5 & 9 in SpecR and CmR</li> | ||
| Line 539: | Line 539: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<ul> | <ul> | ||
| Line 548: | Line 548: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Start midiprep of C-Spec from colony 2 (isopropanol added elute was refrigerated for 4 hrs)</li> | <li>Start midiprep of C-Spec from colony 2 (isopropanol added elute was refrigerated for 4 hrs)</li> | ||
| Line 555: | Line 555: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Dilution of N-Spec and C-Spec (4x)</li> | <li>Dilution of N-Spec and C-Spec (4x)</li> | ||
| Line 567: | Line 567: | ||
</tr> | </tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis of colony PCR</li> | <li>Gel analysis of colony PCR</li> | ||
| Line 578: | Line 578: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Start midiprep of N-Spec (isopropanol added elute refrigerated overnight)</li> | <li>Start midiprep of N-Spec (isopropanol added elute refrigerated overnight)</li> | ||
| Line 586: | Line 586: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Continue midiprep of N-Spec (by ethanol preciptation)</li> | <li>Continue midiprep of N-Spec (by ethanol preciptation)</li> | ||
| Line 593: | Line 593: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Continue midiprep of C-Spec (by ethanol precipitation)</li> | <li>Continue midiprep of C-Spec (by ethanol precipitation)</li> | ||
| Line 602: | Line 602: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Single digest of diluted C-Spec (Eco only)</li> | <li>Single digest of diluted C-Spec (Eco only)</li> | ||
| Line 641: | Line 641: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up digests of N-Spec and C-Spec (Eco and KpnI)- (CD 1)</li> | <li>Set up digests of N-Spec and C-Spec (Eco and KpnI)- (CD 1)</li> | ||
| Line 648: | Line 648: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel purification of N and C Specs from CD 2</li> | <li>Gel purification of N and C Specs from CD 2</li> | ||
| Line 655: | Line 655: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<ul> | <ul> | ||
| Line 663: | Line 663: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Backbone PCR of pSB1C3 using Barns buffer</li> | <li>Backbone PCR of pSB1C3 using Barns buffer</li> | ||
| Line 670: | Line 670: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel purification of pSB1C3</li> | <li>Gel purification of pSB1C3</li> | ||
| Line 679: | Line 679: | ||
</tr> | </tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up digests of N-Spec and C-Spec again, however, with a higher dilution of N-Spec - (CD 2)</li> | <li>Set up digests of N-Spec and C-Spec again, however, with a higher dilution of N-Spec - (CD 2)</li> | ||
| Line 690: | Line 690: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Dephosphorylate vector (C-Spec) from CD 2</li> | <li>Dephosphorylate vector (C-Spec) from CD 2</li> | ||
| Line 698: | Line 698: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation of E.Coli with the two Spec final ligates - C-Spec (vector only) and N and C Specs (vector and insert)</li> | <li>Transformation of E.Coli with the two Spec final ligates - C-Spec (vector only) and N and C Specs (vector and insert)</li> | ||
| Line 705: | Line 705: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Gel analysis and extraction of pSB1C3</li> | <li>Gel analysis and extraction of pSB1C3</li> | ||
| Line 711: | Line 711: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Test digest of final spec minipreps with Eco and Kpn1 and Eco and Spe</li> | <li>Test digest of final spec minipreps with Eco and Kpn1 and Eco and Spe</li> | ||
| Line 750: | Line 750: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up 5 ml culture for miniprep of C-Spec of colnies 1 and 2 from synthesis in AmpR</li> | <li>Set up 5 ml culture for miniprep of C-Spec of colnies 1 and 2 from synthesis in AmpR</li> | ||
| Line 757: | Line 757: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Start midiprep of C-Spec</li> | <li>Start midiprep of C-Spec</li> | ||
| Line 764: | Line 764: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<li>Start midiprep of C-Spec colonies again</li> | <li>Start midiprep of C-Spec colonies again</li> | ||
<ul> | <ul> | ||
| Line 771: | Line 771: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Cloning digests of C-Spec with Kpn1 and Pst and N-Spec with Eco and Kpn1</li> | <li>Cloning digests of C-Spec with Kpn1 and Pst and N-Spec with Eco and Kpn1</li> | ||
| Line 778: | Line 778: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
| Line 790: | Line 790: | ||
</tr> | </tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Miniprep cultures of C-Spec have not grown well enough therefore they will be left overnight</li> | <li>Miniprep cultures of C-Spec have not grown well enough therefore they will be left overnight</li> | ||
| Line 800: | Line 800: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Test digest C-Spec mini with ECo and KpnI</li> | <li>Test digest C-Spec mini with ECo and KpnI</li> | ||
| Line 810: | Line 810: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Continue Midiprep of C-Spec, pellets obtained</li> | <li>Continue Midiprep of C-Spec, pellets obtained</li> | ||
| Line 817: | Line 817: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Ligation gel with inserts; N-Spec (E & Kpn1) and C-Spec (Kpn1 & P) and Vector; pSB1C3 (E & P)</li> | <li>Ligation gel with inserts; N-Spec (E & Kpn1) and C-Spec (Kpn1 & P) and Vector; pSB1C3 (E & P)</li> | ||
| Line 824: | Line 824: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation of E.Coli with the two Spec final ligates - C3 (vector only) and N and C Specs in C3 (vector and insert)</li> | <li>Transformation of E.Coli with the two Spec final ligates - C3 (vector only) and N and C Specs in C3 (vector and insert)</li> | ||
| Line 830: | Line 830: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformations failed, re-try next week!</li> | <li>Transformations failed, re-try next week!</li> | ||
| Line 868: | Line 868: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Set up 5 ml culture for miniprep of C-Spec of colnies 1 and 2 from synthesis in AmpR</li> | <li>Set up 5 ml culture for miniprep of C-Spec of colnies 1 and 2 from synthesis in AmpR</li> | ||
| Line 875: | Line 875: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Start midiprep of C-Spec</li> | <li>Start midiprep of C-Spec</li> | ||
| Line 882: | Line 882: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<li>Start midiprep of C-Spec colonies again</li> | <li>Start midiprep of C-Spec colonies again</li> | ||
<ul> | <ul> | ||
| Line 889: | Line 889: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Cloning digests of C-Spec with Kpn1 and Pst and N-Spec with Eco and Kpn1</li> | <li>Cloning digests of C-Spec with Kpn1 and Pst and N-Spec with Eco and Kpn1</li> | ||
| Line 896: | Line 896: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
| Line 908: | Line 908: | ||
</tr> | </tr> | ||
| - | <td style="background-color:#FFCC66;width:100px;text-align:center;"><b>Afternoon</b> | + | <td style="background-color:#FFCC66;width:100px;text-align:center;color:#555555;"><b>Afternoon</b> |
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Miniprep cultures of C-Spec have not grown well enough therefore they will be left overnight</li> | <li>Miniprep cultures of C-Spec have not grown well enough therefore they will be left overnight</li> | ||
| Line 918: | Line 918: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Test digest C-Spec mini with ECo and KpnI</li> | <li>Test digest C-Spec mini with ECo and KpnI</li> | ||
| Line 928: | Line 928: | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Continue Midiprep of C-Spec, pellets obtained</li> | <li>Continue Midiprep of C-Spec, pellets obtained</li> | ||
| Line 935: | Line 935: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Ligation gel with inserts; N-Spec (E & Kpn1) and C-Spec (Kpn1 & P) and Vector; pSB1C3 (E & P)</li> | <li>Ligation gel with inserts; N-Spec (E & Kpn1) and C-Spec (Kpn1 & P) and Vector; pSB1C3 (E & P)</li> | ||
| Line 942: | Line 942: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformation of E.Coli with the two Spec final ligates - C3 (vector only) and N and C Specs in C3 (vector and insert)</li> | <li>Transformation of E.Coli with the two Spec final ligates - C3 (vector only) and N and C Specs in C3 (vector and insert)</li> | ||
| Line 948: | Line 948: | ||
</td> | </td> | ||
| - | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;"> | + | <td style="background-color:#e7e7e7;height:100px;width:200px;text-align:left;color:#555555;"> |
<ul> | <ul> | ||
<li>Transformations failed, re-try next week!</li> | <li>Transformations failed, re-try next week!</li> | ||
Revision as of 23:22, 27 October 2010
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
| Objectives |
|
We are assembling the AmyE and PyrD vectors in order to transform B. Subtilis with our parts. Once completed, these vectors will be reusable and can then be used to introduce any relevant piece of DNA directly into B.subtilis genome. The vectors will be used both for the final assembly and for testing constructs. |
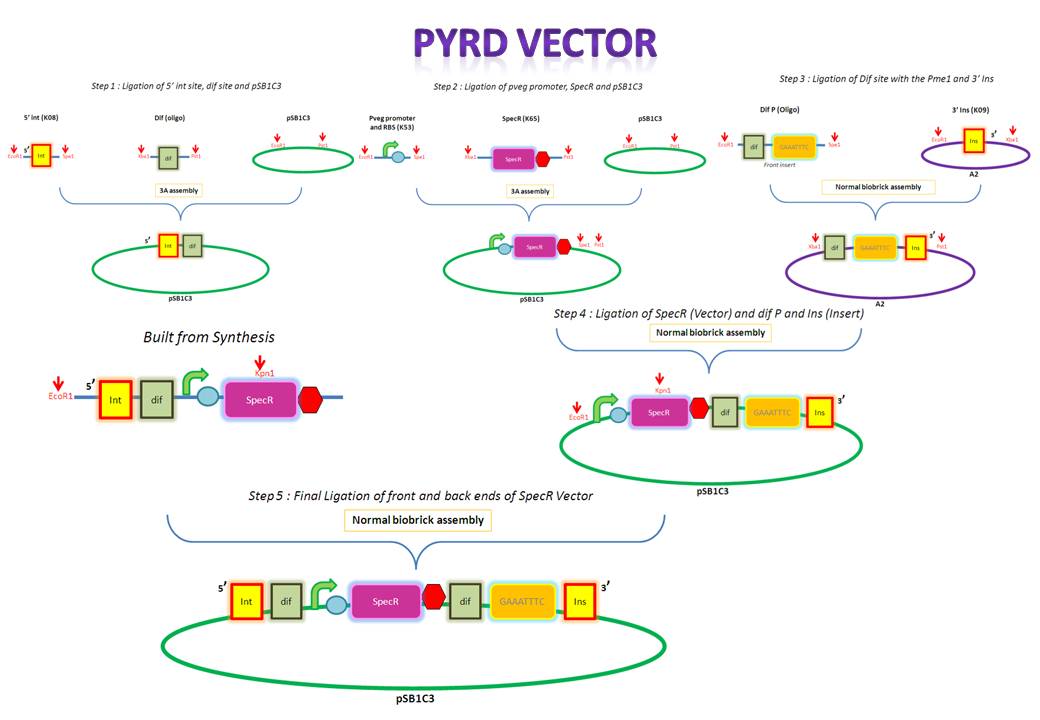
| PyrD Vector |
| Week 6 | |||||||||||||||||||||
|
Thursday, August 12
The forward and reverse strands of the 5' dif site with XbaI and PstI restriction sites on either side have been synthesized separately. The synthesized fragments arrive in solid powder form. These were immediately diluted in ddH2O to obtain a stock concentration of 1 ng/ul. They were then allowed to anneal together by first heating them to 95 degs for denaturation and allowing them to cool down and anneal overnight. Friday, August 13
After annealing the two strands, the oligo was cut with XbaI and PstI to obtain overhangs that would later ligate with the compatible overhangs of a cut vector.
The cut oligo was PCR purified in order to get rid of any contaminants. PCR purification gets rid of short pieces of DNA which are less than about 40 base pairs.
|
| Week 7 | ||||||||||||||||||
|
K143008 is the 5' integration site for the PyrD vector. This will be used as a front insert together with the dif XP for the pSB1C3 vector.
The pSB1C3 vector backbone from the registry was amplified with the use of SB3 and SB2a primers. Submission of parts to the registry requires them to be in a pSB1C3 vector therefore any parts to be submitted will be inserted into this vector.
The PCR amplified vector was purified in order to get rid of any contaminants. For example, short pieces of DNA like the primers. Tuesday, August 17
The digested 5' ins was first analyzed on the gel to verify it's size and then extracted for purification. The 5' ins was gel purified in order to extract only the relevant piece of DNA.
The pSB1C3 vector was digested so that it would contain compatible overhangs for ligation with inserts.
The digested pSB1C3 was re-purified in order to get rid of any contaminant DNA that arose during the digestion.
pveg (promoter and RBS) and Spec-T (Spectinomycin with a terminator) were digested in preparation for 3A assembly.
The digested 5' ins and dif (the front inserts) were ligated overnight with pSB1C3 (the vector). A bench ligation and an overnight ligation were set up.
E.Coli was transformed via the chemical method using the bench ligate.
Since these are both inserts they were gel extracted and purified. PCR purification is not carried out for inserts since they are small and would therefore be lost during the process. Wednesday, August 18
pveg and SpecR-T (the front inserts) were ligated overnight with pSB1C3 (the vector). Thursday, August 19
Friday, August 20
The forward and reverse strands of the 3' dif Pme1 sites with XbaI and PstI restriction sites on either side have been synthesized separately. The synthesized fragments arrive in solid powder form. These were immediately diluted in ddH2O to obtain a stock concentration of 1 ng/ul. They were then allowed to anneal together by heating them to 95 degs for denaturation and allowing them to cool down and anneal overnight. |
| Week 8 | |||||||||||||||||||||
|
|
| Week 9 | ||||||||||||||||||
|
|
| Week 10 | ||||||||||||||||||
|
|
| Week 11 | ||||||||||||||||||
|
|
| Week 12 | |||||||||||||||||||||
|
|
| Week 13 | |||||||||||||||||||||
|
|
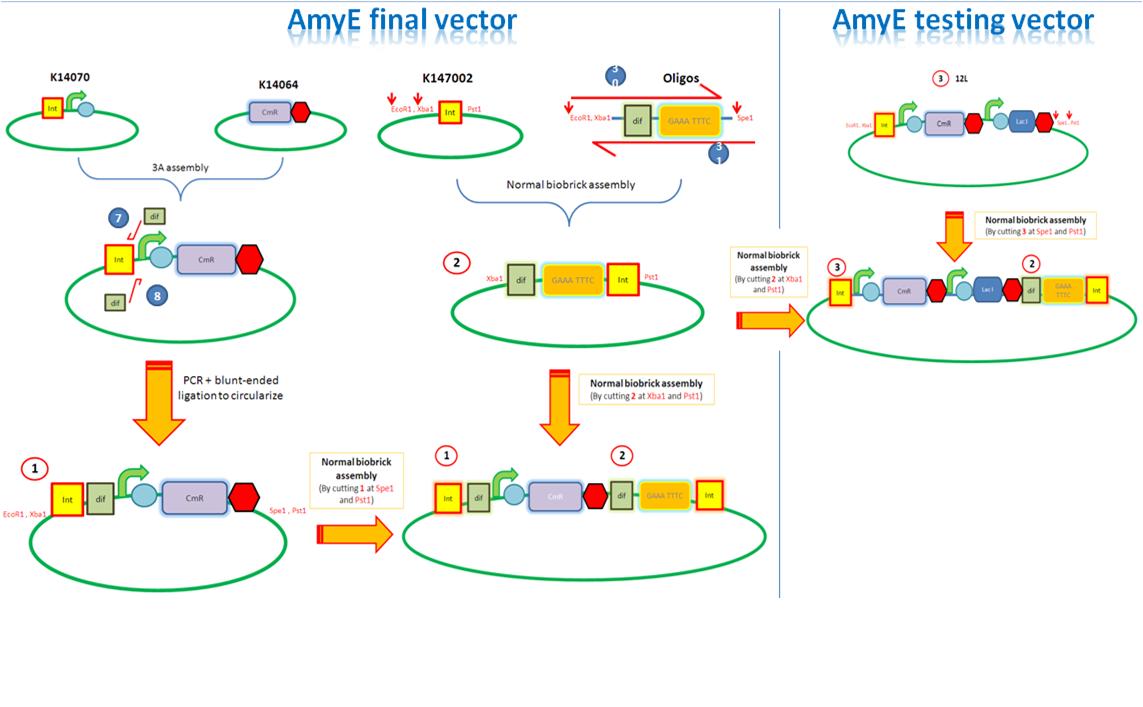
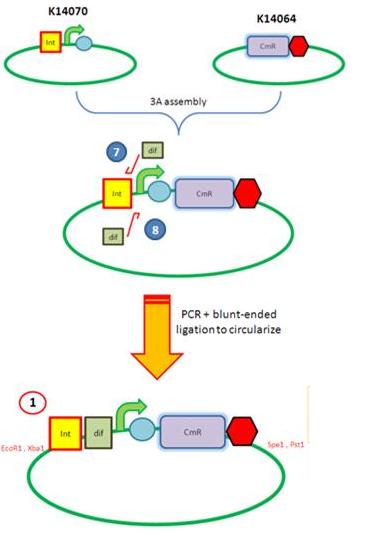
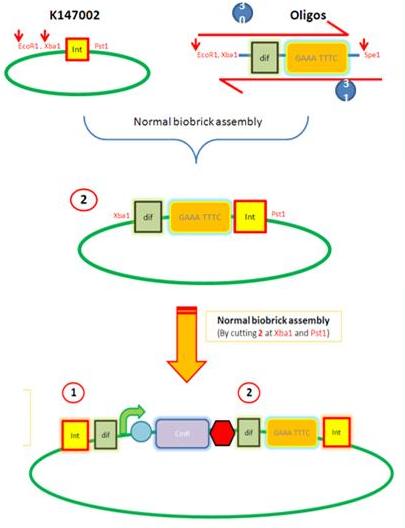
| AmyE Vector |
|
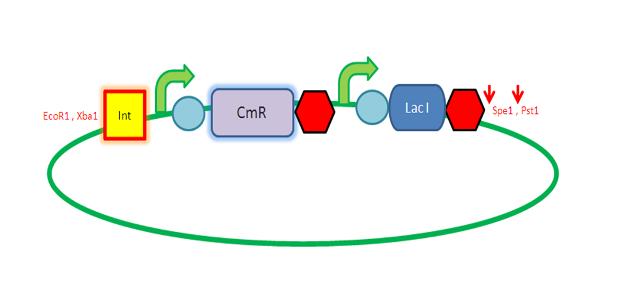
Starting from the top, we are assembling the first two fragments (K14070 and K14064) and K147002 with our oligos to add in a Dif site. Two dif sites on either side of resistance cassettes can be used to later excise antibiotic resistance from our final constructs.
Next Steps:
K14002 and oligos
Next Steps:
|
| Schedule & Lab Notes |
| Week 7 | ||||||||||||||||||
|
|
| Week 8 | |||||||||||||||||||||
|
|
| Week 9 | ||||||||||||||||||
|
|
 "
"