Team:TU Munich/Parts
From 2010.igem.org
(→Part Overview) |
Hartlmueller (Talk | contribs) |
||
| Line 7: | Line 7: | ||
| - | = | + | =Parts Overview= |
| + | ==Submitted Parts== | ||
<groupparts align:"center">igem2010 TU_Munich</groupparts> | <groupparts align:"center">igem2010 TU_Munich</groupparts> | ||
| - | + | == | |
| - | + | ==Parts Falsification and Verification== | |
| - | + | ||
| - | === | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | |
<br> | <br> | ||
{| class="bordertable" width=100% align="center" style="text-align: center; font-size:80%" | {| class="bordertable" width=100% align="center" style="text-align: center; font-size:80%" | ||
| Line 151: | Line 108: | ||
| Basic plasmid backbone with kanamycin resistance | | Basic plasmid backbone with kanamycin resistance | ||
| style="background:#4e9d20"|worked | | style="background:#4e9d20"|worked | ||
| + | | existing | ||
| + | |||
| + | |- align=”center” | ||
| + | | <partinfo>pSB1A10</partinfo> | ||
| + | | BioBrick backbone for measuring termination efficiency | ||
| + | | Plasmid Backbone | ||
| + | | Plasmid backbone with ampicillin resistance (based on pSB1A2) | ||
| + | | style="background:red"|failed | ||
| existing | | existing | ||
|} | |} | ||
<br> | <br> | ||
| + | |||
| + | |||
| + | |||
| + | =Parts Description= | ||
| + | |||
| + | ==Malachite Green Binding Aptamer - BBa_K494000== | ||
| + | <br> | ||
| + | Methods to visualize nucleic acids via fluorescence are rare, partly due to the size of fluorescent reporters. Thus, we present the malachite green-binding aptamer to the partsregistry. By adding only 38 bp, fluorescent determination of specific nucleic acids becomes possible allowing evalutation of PoPS-based devices via in vitro transcription.<sup>[[Team:TU_Munich/Parts#ref1|[1]]]</sup> Binding of triphenyl dye malachite green to the aptamer increases fluorescence by 2360-fold. This leads to an significant increase and a shift in absorbance from 618 to 630 nm. With an emission maximum at 652 nm, aptamer-bound malachite green fluoresces at longer wavelength than most dyes and does not interfere with those.<sup>[[Team:TU_Munich/Parts#ref2|[2]]]</sup> We provide this part for efficient ''in vitro'' evaluation of PoPS-based devices in general and switches based on our concept in particular. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | The malachite green binding aptamer has been successfully used in screening systems being both robust and easy to produce. Aptamers provide specifities in the range of antibodies and can be evolved to target small molecules and proteins.<sup>[[Team:TU_Munich/Parts#ref3|[3]]]</sup> <br> | ||
| + | Since malachite green is a membrane permeable dye, its uses are not limited to ''in vitro'' measurements. The malachite green aptamer can be used to tag and follow any RNA, including messengar and small RNAs to study questions about their metabolism and biological functions.<sup>[[Team:TU_Munich/Parts#ref2|[2]]]</sup> Aside from the application as a mere reporter, the malachite green binding aptamer has already been utilized to build up modular sensors which can together with another RNA-binding domain sense and report small molecules like ATP for example. This new detection method seems to provide promising future applications and sensors.<sup>[[Team:TU_Munich/Parts#ref4|[4]]]</sup> Since the principle of modularizing fits well into our concept of building networks, we like to provide this part to allow further engineering considering ''in vitro'' sensing systems. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ==Plasmids== | ||
| + | In general we want to provide a new principle of gene regulation which can be further developed, tested and optimizted by everybody. Therefore we focus on providing the parts needed for verification and testing of new individual switches. We provide a plasmid which can be used for further cloning, a positive control to test the general functionality and the constructs we characterized for comparison. | ||
| + | |||
| + | ===Screening system: Backbone BBa_K494001=== | ||
| + | We designed a new screening systems based on the non-functional pSB1A10 plasmid. We improved its features for ‘’in vivo’’ characterization of PoPS-based devices using fluorescent reporters . The plasmid still contains the Pbad arabinose-inducible induction system as a tunable input and eGFP as an internal standard for induction. However, we altered the reporter protein to mCherry. Furthermore we adjusted the BioBrick cloning site to allow cloning of additional parts independent from the Input/Output measurement. This screening plasmid is designed to be used with a second Arabinose inducible promoter <partinfo>I13453</partinfo> which is not included in this part. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | The improved screening system is optimized for the evaluation of PoPS-based devices in fluorescence measurements. RFP which was known to contain an RNase restriction site was replaced by mCherry which combines good expression yield, short maturation times and an acceptable and well-characterized quantum yield. A unique challenge for the characterization of our switches is the expression of a corresponding signal independent of the input/output measurement. Thus, we moved the BioBrick cloning site resulting in the fluorescent reporter being inside the cloning site and giving the possibility to clone independent parts behind the reporter protein. | ||
| + | To fully function our screening plasmid need the arabinose inducible promoter BBa_I13453 in front of the PoPS-based device to screen. Using a second arabinose inducible promoter, we were able to keep eGFP as an internal standard for the tunable input via the Pbad arabinose-inducible induction system. The two identical promoters ensure the same rate of induction for eGFP and the tested PoPS-based device. Thus, obtaining comparable screening results is easy. Unfortunately, this design implicates a minor disadvantage. Two cloning steps are needed to gain an functional construct for testing any PoPS-based device. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ===Screening System: Positive Control BBa_K494002=== | ||
| + | [[Image:TUM2010_PicPosControl3cInd.png|70px|left|]] | ||
| + | Here we present a ready to use composite part consisting of our screening system pSB1A10mod <partinfo>BBa_K494001</partinfo>, the PBad Promotor <partinfo>BBa_I13453</partinfo> and the reporter protein mCherry <partinfo>BBa_J06702</partinfo> including RBS and double Terminator <partinfo>BBa_B0015</partinfo>. This part belongs to the screening system and is intended as positive control.<br> | ||
| + | <br> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | [[Image:TUM2010_graphPosControl3cInd.png|400px|thumb|center|Emission spectra of induced pSB1A10mod positive control BBa_K494002 ; A: eGFP fluorescence ex: 501 nm, B: mCherry fluorescence ex: 587 nm]] | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ===Screening System: Negative Control BBa_K494003=== | ||
| + | Here we present a ready to use composite part consisting of our screening system pSB1A10mod BBa_K494001, the PBad Promotor BBa_I13453, the His Terminator and the reporter protein mCherry BBa_J06702 including RBS and double Terminator BBa_B0015. This part belongs to the screening system and is intended as negative control. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | [[Image:TUM2010_HisTermInd.png|400px|thumb|center|Emission spectra of induced pSB1A10mod HisTerm BBa_K494004 ; A: eGFP fluorescence ex: 501 nm, B: mCherry fluorescence ex: 587 nm]] | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | ===Screening System: Control BBa_K494004=== | ||
| + | Here we present a ready to use composite part consisting of our screening system pSB1A10mod BBa_K494001, the PBad Promotor BBa_I13453 and the reporter protein mCherry BBa_J06702 including RBS and double Terminator BBa_B0015. This part belongs to the screening system and is intended as control for light termination. | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| + | [[Image:TUM2010_graphTrpTermInd.png|400px|thumb|center|Emission spectra of induced pSB1A10mod TrpTerm BBa_K494004 ; A: eGFP fluorescence ex: 501 nm, B: mCherry fluorescence ex: 587 nm]] | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
| + | === Screening System: BBa_K494005 === | ||
| + | |||
| + | === Screening System: BBa_K494006 === | ||
| + | |||
=Falsified Parts= | =Falsified Parts= | ||
Revision as of 12:17, 27 October 2010
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Parts OverviewSubmitted Parts<groupparts align:"center">igem2010 TU_Munich</groupparts> == Parts Falsification and Verification
Parts DescriptionMalachite Green Binding Aptamer - BBa_K494000
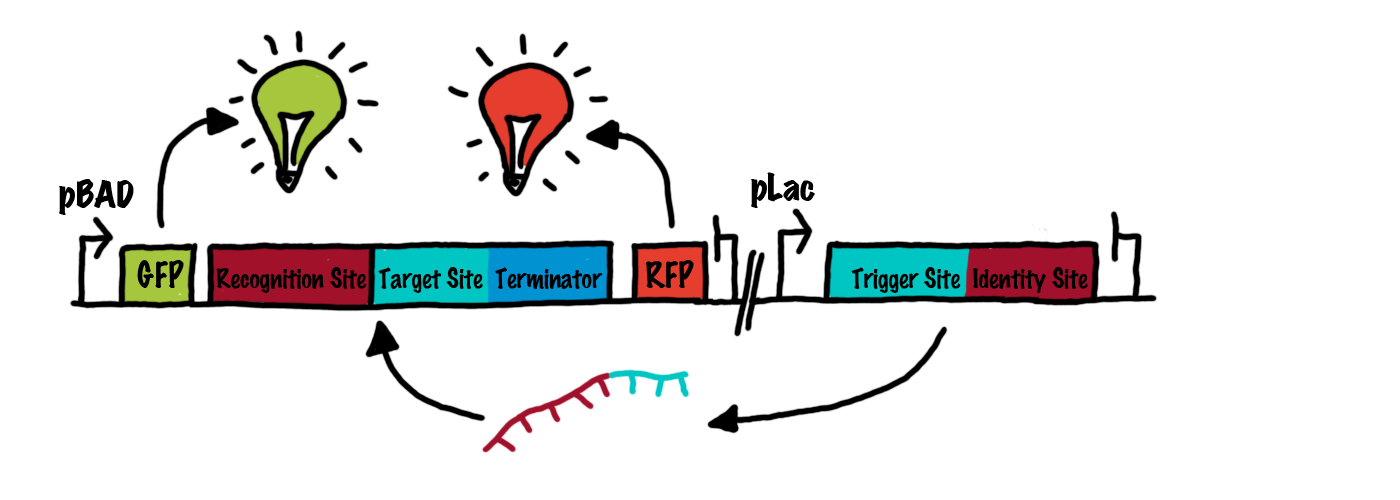
The malachite green binding aptamer has been successfully used in screening systems being both robust and easy to produce. Aptamers provide specifities in the range of antibodies and can be evolved to target small molecules and proteins.[3] PlasmidsIn general we want to provide a new principle of gene regulation which can be further developed, tested and optimizted by everybody. Therefore we focus on providing the parts needed for verification and testing of new individual switches. We provide a plasmid which can be used for further cloning, a positive control to test the general functionality and the constructs we characterized for comparison. Screening system: Backbone BBa_K494001We designed a new screening systems based on the non-functional pSB1A10 plasmid. We improved its features for ‘’in vivo’’ characterization of PoPS-based devices using fluorescent reporters . The plasmid still contains the Pbad arabinose-inducible induction system as a tunable input and eGFP as an internal standard for induction. However, we altered the reporter protein to mCherry. Furthermore we adjusted the BioBrick cloning site to allow cloning of additional parts independent from the Input/Output measurement. This screening plasmid is designed to be used with a second Arabinose inducible promoter <partinfo>I13453</partinfo> which is not included in this part. The improved screening system is optimized for the evaluation of PoPS-based devices in fluorescence measurements. RFP which was known to contain an RNase restriction site was replaced by mCherry which combines good expression yield, short maturation times and an acceptable and well-characterized quantum yield. A unique challenge for the characterization of our switches is the expression of a corresponding signal independent of the input/output measurement. Thus, we moved the BioBrick cloning site resulting in the fluorescent reporter being inside the cloning site and giving the possibility to clone independent parts behind the reporter protein. To fully function our screening plasmid need the arabinose inducible promoter BBa_I13453 in front of the PoPS-based device to screen. Using a second arabinose inducible promoter, we were able to keep eGFP as an internal standard for the tunable input via the Pbad arabinose-inducible induction system. The two identical promoters ensure the same rate of induction for eGFP and the tested PoPS-based device. Thus, obtaining comparable screening results is easy. Unfortunately, this design implicates a minor disadvantage. Two cloning steps are needed to gain an functional construct for testing any PoPS-based device. CloseScreening System: Positive Control BBa_K494002Here we present a ready to use composite part consisting of our screening system pSB1A10mod <partinfo>BBa_K494001</partinfo>, the PBad Promotor <partinfo>BBa_I13453</partinfo> and the reporter protein mCherry <partinfo>BBa_J06702</partinfo> including RBS and double Terminator <partinfo>BBa_B0015</partinfo>. This part belongs to the screening system and is intended as positive control. Screening System: Negative Control BBa_K494003Here we present a ready to use composite part consisting of our screening system pSB1A10mod BBa_K494001, the PBad Promotor BBa_I13453, the His Terminator and the reporter protein mCherry BBa_J06702 including RBS and double Terminator BBa_B0015. This part belongs to the screening system and is intended as negative control. Screening System: Control BBa_K494004Here we present a ready to use composite part consisting of our screening system pSB1A10mod BBa_K494001, the PBad Promotor BBa_I13453 and the reporter protein mCherry BBa_J06702 including RBS and double Terminator BBa_B0015. This part belongs to the screening system and is intended as control for light termination. Screening System: BBa_K494005Screening System: BBa_K494006Falsified PartspSB1A10The Screening Plasmid pSB1A10 is intended for the characterization of an Input/Output for a PoPS based device on the basis of two fluorescent proteins. While the first fluorescent protein, eGFP quantifies the input from an arabinose induced promoter, the second reporter, RFP detects the output. However, our experiments designed for testing toggle switches revealed the part to be non-functional. In the current available form, RFP fluorescence is not induced even when the PoPS device does not interfere with the Polymerase.
Although induction with arabinose worked fine indicated by GFP fluorescence, we could not detect any RFP output. The kinetic measurment on the left shows pSB1A10 postive control with a nonsense sequence inserted into the BioBrick site between the two fluorescent proteins GFP and RFP. Nevertheless induction does not trigger any RFP expression. Therefore no output signal is created in our experimantal setup. This fact renders the part as it is unsuitable for testing PoPS-based devices. We first assumed degradation of RFP-mRNA which is known to have an RNase site might be responsible. But comparison with our new screening system reveals a transcriptional problem to be most likely as replacing RFP did not improve the construct. Though, introducing a second arabinose promoter finally resulted in the desired output signal.
References[1] Grate, D. and C. Wilson, Laser-mediated, site-specific inactivation of RNA transcripts. Proceedings of the National Academy of Sciences of the United States of America, 1999. 96(11): p. 6131. [2] Babendure, J.R., S.R. Adams, and R.Y. Tsien, Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc, 2003. 125(48): p. 14716-14717. [3] Rowe, W., M. Platt, and P.J.R. Day, Advances and perspectives in aptamer arrays. Integrative Biology, 2009. 1(1): p. 53-58. [4] Stojanovic, M.N. and D.M. Kolpashchikov, Modular aptameric sensors. J. Am. Chem. Soc, 2004. 126(30): p. 9266-9270. [5] Smolke and so on.... [6] http://en.wikipedia.org/wiki/Logic_gate#Symbols |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"