Team:Washington/Tools Created/New Software
From 2010.igem.org
(→A Standard for Electronic Diagrams) |
(→BBF RFC 68: A Standard for Electronic Diagrams) |
||
| Line 29: | Line 29: | ||
====<center>BBF RFC 68: A Standard for Electronic Diagrams</center>==== | ====<center>BBF RFC 68: A Standard for Electronic Diagrams</center>==== | ||
| - | Distribution of standardized diagrams describing DNA construct designs for synthetic biology depends both on the use of a standard set of symbols and on the method of distribution. Therefore the two main requirements for our propoesed standard, | + | Distribution of standardized diagrams describing DNA construct designs for synthetic biology depends both on the use of a standard set of symbols and on the method of distribution. Therefore the two main requirements for our propoesed standard, [http://openwetware.org/wiki/The_BioBricks_Foundation:RFC#BBF_RFC_68:_Standard_for_the_Electronic_Distribution_of_SBOLv_Diagrams|BBF RFC 68], are: |
# The use of SBOLv symbols | # The use of SBOLv symbols | ||
# The linking of symbols to reference entry pages on the Registry of Standard Biological Parts using http URLs | # The linking of symbols to reference entry pages on the Registry of Standard Biological Parts using http URLs | ||
Revision as of 04:53, 27 October 2010
As synthetic biologists create ever more complex systems, automation and computer-aided design are becoming indispensable. Computational tools will eventually play a central role in the design, construction, and testing of new devices. To meet the emerging needs of the synthetic biology community, the University of Washington 2010 iGEM team has developed a standard for the electronic distribution of diagrams (BBF RFC 68), as well as two new software tools, WikiDust and PartsRobot.
BBF RFC 68: A Standard for Electronic Diagrams
Distribution of standardized diagrams describing DNA construct designs for synthetic biology depends both on the use of a standard set of symbols and on the method of distribution. Therefore the two main requirements for our propoesed standard, [http://openwetware.org/wiki/The_BioBricks_Foundation:RFC#BBF_RFC_68:_Standard_for_the_Electronic_Distribution_of_SBOLv_Diagrams|BBF RFC 68], are:
- The use of SBOLv symbols
- The linking of symbols to reference entry pages on the Registry of Standard Biological Parts using http URLs
The result constitutes a diagram which allows the reader to click on portions of the diagram which correspond to SBOLv symbols and read the reference information about that particular portion of the diagram.
WikiDust is a plugin for TinkerCell, a computer-aided design tool developed here at the [http://www.washington.edu/ University of Washington]. WikiDust can be used to export diagrams of TinkerCell models to the iGEM wiki or other webpages. Our goal in creating WikiDust is to provide an easy way to share models and to encourage iGEM teams and researchers to describe their parts more completely. This, in turn, will help others reuse the devices in the future projects.
The first thing WikiDust does is provide a method to associate TinkerCell items with parts on the registry. The same mechanism could also be extended in the future to download reaction kinetics and other parameters from sources like the [http://www.ncbi.nlm.nih.gov/pubmed/20160009 CellML Model Repository] or the [http://www.ncbi.nlm.nih.gov/pubmed/16381960 Biomodels Database]. For now though, it demonstrates the feasability of using a [http://www.sbolstandard.org publicly available semantic knowledgebase] to retrieve part information.
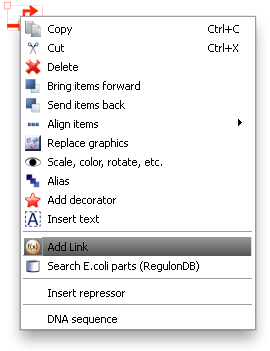
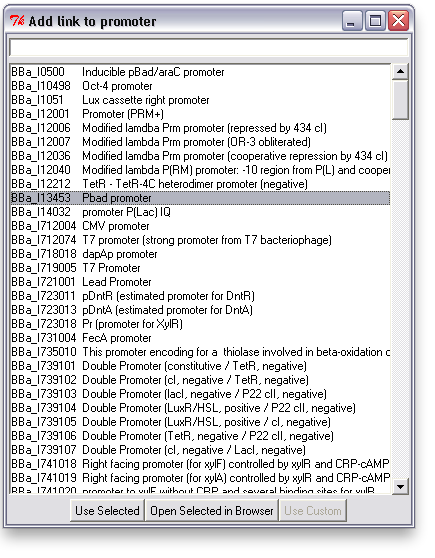
To associate a link with an item in TinkerCell, right click on it and choose "Add Link". This will bring up a search window with suggested parts. You can search for a phrase, a BioBrick ID, etc. You can also open parts in a browser to read more about them or confirm that you've found the right one.
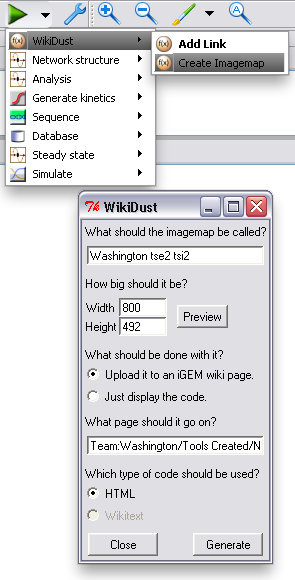
After the model of your device or system is finished, and you've added all the links you want, you can upload a representation of it to your page. Just choose "Create Imagemap" from the TinkerCell plugins menu, and change any options you want in the window that pops up. WikiDust will automatically handle uploading to the iGEM wiki, or it can just display code for you to copy and paste to another site. Clicking an item in the uploaded diagram will go to the correct part page. In fact, our Tse2/Tsi2 diagram was generated using WikiDust.
To start using WikiDust, you’ll need the main TinkerCell program, as well as Python and Subversion. All three of these can be found on the [http://www.tinkercell.com/downloads-2 TinkerCell download page].
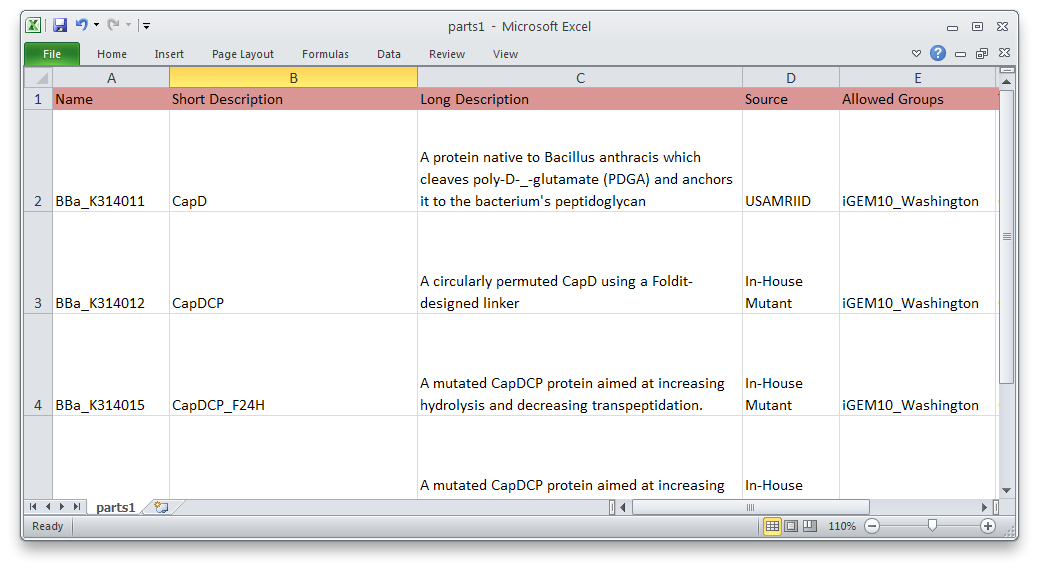
Part Submission Made Easy
We have also developed a command-line tool to submit multiple parts to the [http://partsregistry.org/Main_Page Registry] at once. This will make it much easier and faster to submit a series of similar parts—promoters of different strengths, variations on a protein, etc. We used it to submit most of our parts.
Using PartsRobot is simple. Just fill out the template spreadsheet with information about each part. Then, in a command prompt, type:
python partsrobot.py submit yourspreadsheet.csv
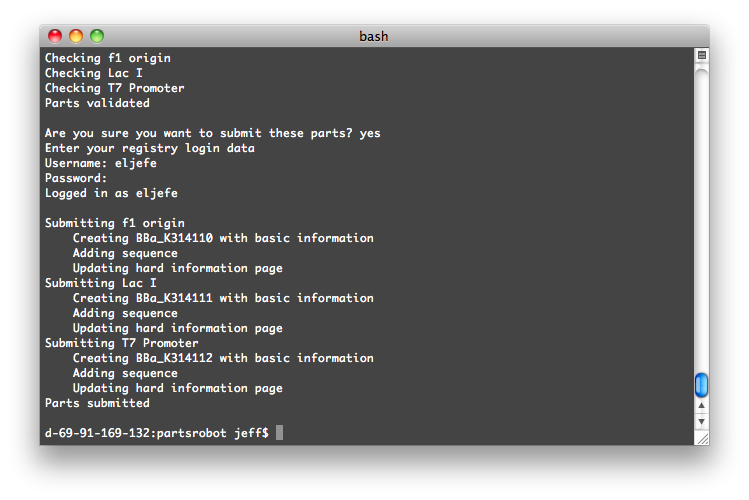
The program will log into the Registry and check your parts for syntax errors. Then, after you give the go-ahead, it will submit them.
Get PartsRobot from the [http://sourceforge.net/p/partsrobot/home/ SourceForge page].
 "
"