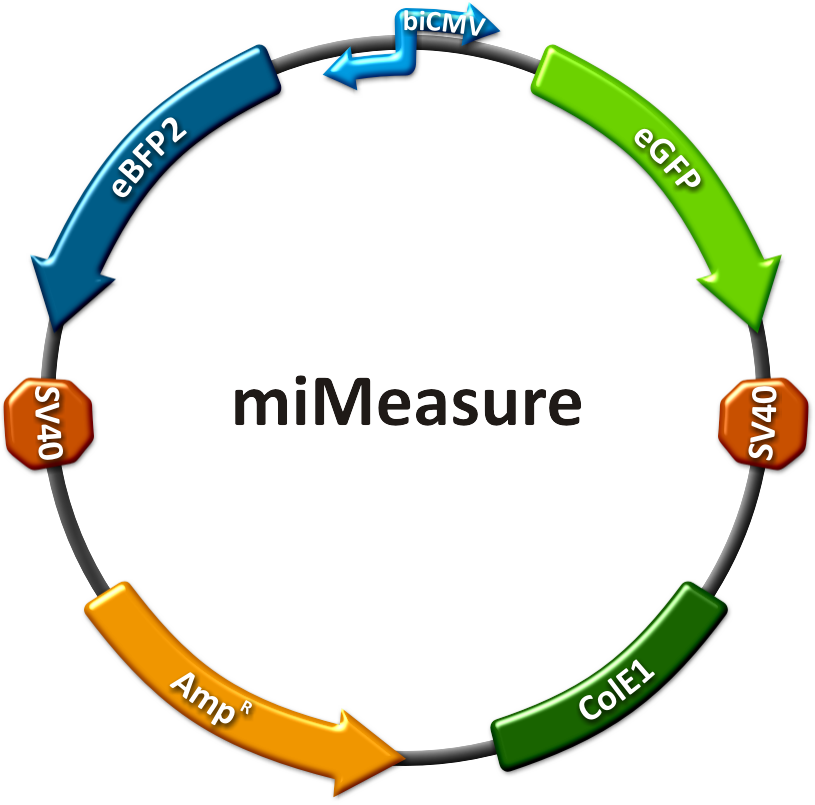
Team:Heidelberg/Project/miMeasure
From 2010.igem.org
(→Analysis of raPCR generated binding sites) |
(→miMeasure) |
||
| Line 34: | Line 34: | ||
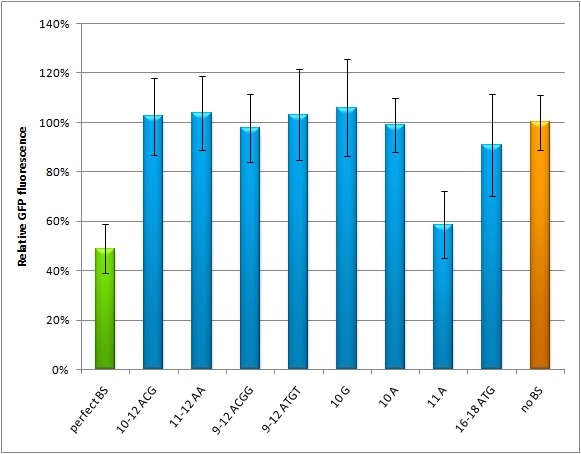
The raPCR generates binding sites, which are aligned by chance with each other, whereby different random spacer regions are inserted in between. If perfect binding sites are aligned, this is predicted to give a stronger knock-down than a single one. If imperfect binding sites are aligned, it is also supposed to be stronger than a single one. In this way binding sites with different knock-down efficiencies can be generated. | The raPCR generates binding sites, which are aligned by chance with each other, whereby different random spacer regions are inserted in between. If perfect binding sites are aligned, this is predicted to give a stronger knock-down than a single one. If imperfect binding sites are aligned, it is also supposed to be stronger than a single one. In this way binding sites with different knock-down efficiencies can be generated. | ||
| - | This experiment shows, that the binding site pattern with 3 aligned perfect binding sites (miM-1.3-7) gives the strongest knock-down. Whereby the binding site pattern with two imperfect binding sites (miM-3.1-8) is weaker, but still stronger than the negative control. If one binding site is randomized from nucleotide 9-12 or 9-22 (miM-r12, miM-r22), it loses its capability of protein down-regulation. | + | This experiment shows, that the binding site pattern with 3 aligned perfect binding sites ([https://2010.igem.org/Team:Heidelberg/Notebook/miMeasure#Table2 miM-1.3-7]) gives the strongest knock-down. Whereby the binding site pattern with two imperfect binding sites (miM-3.1-8) is weaker, but still stronger than the negative control. If one binding site is randomized from nucleotide 9-12 or 9-22 (miM-r12, miM-r22), it loses its capability of protein down-regulation. |
==Discussion== | ==Discussion== | ||
Revision as of 23:33, 26 October 2010

|
|
||
 "
"