Team:Heidelberg/Project/miMeasure
From 2010.igem.org
(→Analysis of mutated/ randomized binding sites) |
(→Abstract) |
||
| Line 5: | Line 5: | ||
==Abstract== | ==Abstract== | ||
| - | With the | + | With the rising importance of small RNA molecules in gene therapy the identification and characterization of miRNAs and their binding sites become crucial for innovative applications. In order to exploit the miRNA ability to target and regulate specific genes, we constructed a measurement standard (see sidebar) not only to characterize existing miRNAs but also to validate potential synthetic shRNA miRNAs for a new therapeutic approach. The synthetic miRNAs we created lack endogenous targets and are thus applicable for gene regulation without any side effects. This openes new possiblities of precise expression tuning. |
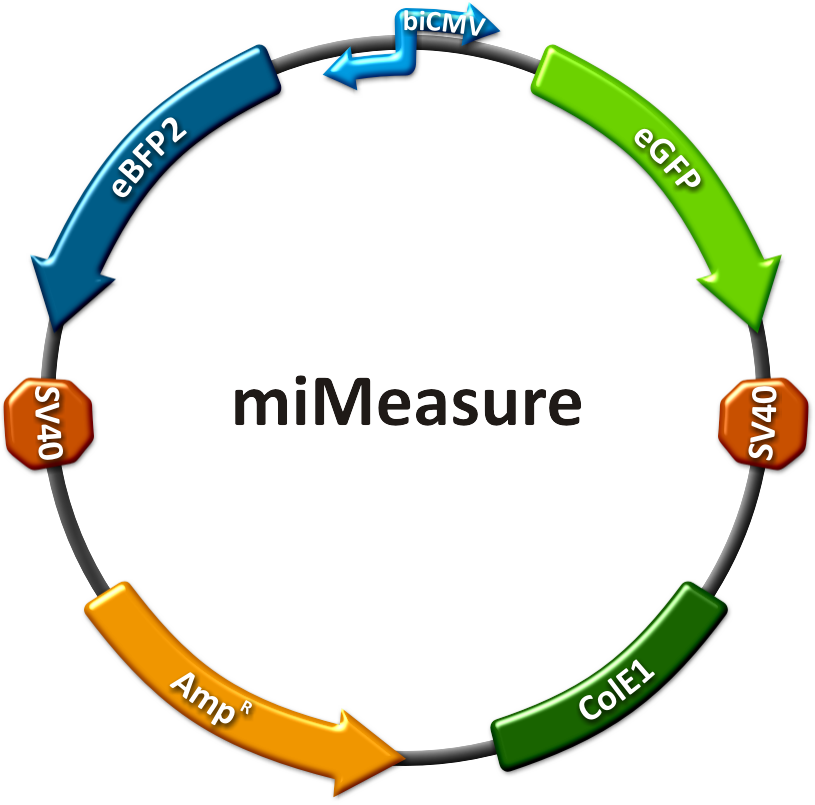
Our miMeasure plasmid normalizes knockdown of the green fluorescent protein (EGFP) to the blue fluorescent protein (EBFP2). This allows an accurate study of binding site properties, since both fluorescent proteins are combined in the same construct and driven by the same bidirectional promoter. Another advantage is, that any desired binding site can be cloned easily into the miMeasure plasmid with the BB_2 standard. As the binding site is inserted downstream of EGFP, a regulation of EGFP expression is to be expected. | Our miMeasure plasmid normalizes knockdown of the green fluorescent protein (EGFP) to the blue fluorescent protein (EBFP2). This allows an accurate study of binding site properties, since both fluorescent proteins are combined in the same construct and driven by the same bidirectional promoter. Another advantage is, that any desired binding site can be cloned easily into the miMeasure plasmid with the BB_2 standard. As the binding site is inserted downstream of EGFP, a regulation of EGFP expression is to be expected. | ||
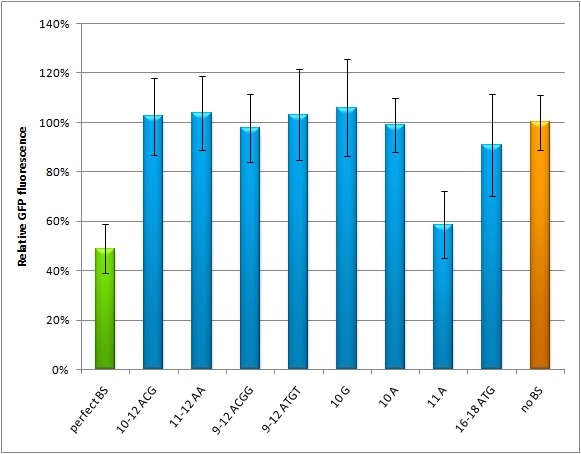
The percentage of knockdown of each modified binding site can be determined by the comparison of the ratio of EGFP to EBFP2 ratio compared to the ratio of the perfect binding site caused knock-down. The ratio is derived from a linear regression curve. Therefore the knock-down efficiency can be conducted by various basic methods e.g. plate reading, flow cytometry or microscopy. | The percentage of knockdown of each modified binding site can be determined by the comparison of the ratio of EGFP to EBFP2 ratio compared to the ratio of the perfect binding site caused knock-down. The ratio is derived from a linear regression curve. Therefore the knock-down efficiency can be conducted by various basic methods e.g. plate reading, flow cytometry or microscopy. | ||
| - | |||
==Introduction== | ==Introduction== | ||
Revision as of 22:16, 26 October 2010

|
|
||
 "
"