Team:Heidelberg/Project/miMeasure
From 2010.igem.org
(→Analysis of mutated/ randomized binding sites) |
(→Analysis of raPCR generated binding sites) |
||
| Line 33: | Line 33: | ||
=====Analysis of raPCR generated binding sites===== | =====Analysis of raPCR generated binding sites===== | ||
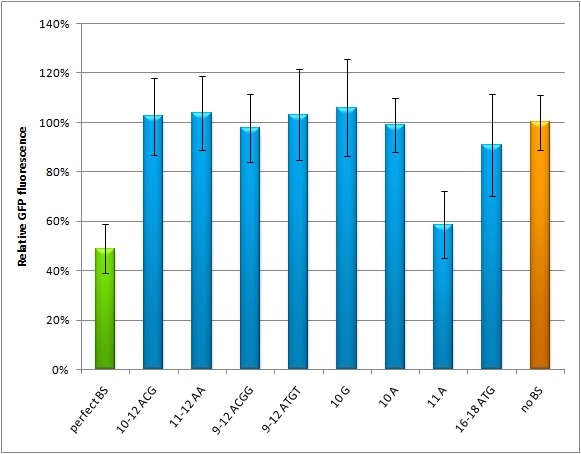
| - | + | The raPCR generates binding sites, which are aligned by chance with each other with different random spacer regions. If perfect binding sites are aligned, this is predicted to give a stronger knock-down than a single one. If imperfect binding sites are aligned, it is also supposed to be stronger than a single one. In this way binding sites with different knock-down efficiencies can be generated. | |
| - | + | This experiment shows, that the binding site pattern with 3 aligned perfect binding sites gives the strongest knock-down. | |
==Discussion== | ==Discussion== | ||
Revision as of 21:48, 26 October 2010

|
|
||
 "
"