Team:Imperial College London/Modelling
From 2010.igem.org
(Difference between revisions)
(signalling model) |
m |
||
| Line 96: | Line 96: | ||
<b>Signalling Modelling Model</b><br/> | <b>Signalling Modelling Model</b><br/> | ||
| + | Goals: | ||
| + | The aim of this model is to determine under which conditions the signalling transduction will happen in our bacteria. | ||
| + | |||
| + | Elements of the system: | ||
| + | <ol> | ||
| + | <li>The signalling model consists of ComD and ComE which are expressed by our bacteria, as well as AIP and Phosphate. These species are all assumed to be present in the cell at a sufficient concentration.</li> | ||
| + | <li>The ComD receptor is activated by the AIP. This triggeres phosphorylation of the ComD receptor. The Phosphate group of the ComD receptor then binds to ComE. The phosphorylated ComE binds to the DNA and acts as a transcription factor.</li> | ||
| + | </ol> | ||
| + | |||
| + | Major assumptions: | ||
<ol> | <ol> | ||
<li>ComD and ComE are present in the cell/cell wall at a high concentration. ComD and ComE are both in steady-state, so the production and degradation constants are negligible.</li> | <li>ComD and ComE are present in the cell/cell wall at a high concentration. ComD and ComE are both in steady-state, so the production and degradation constants are negligible.</li> | ||
Revision as of 15:38, 25 October 2010
| Modelling | Overview | Detection Model | Signaling Model | Fast Response Model | Interactions |
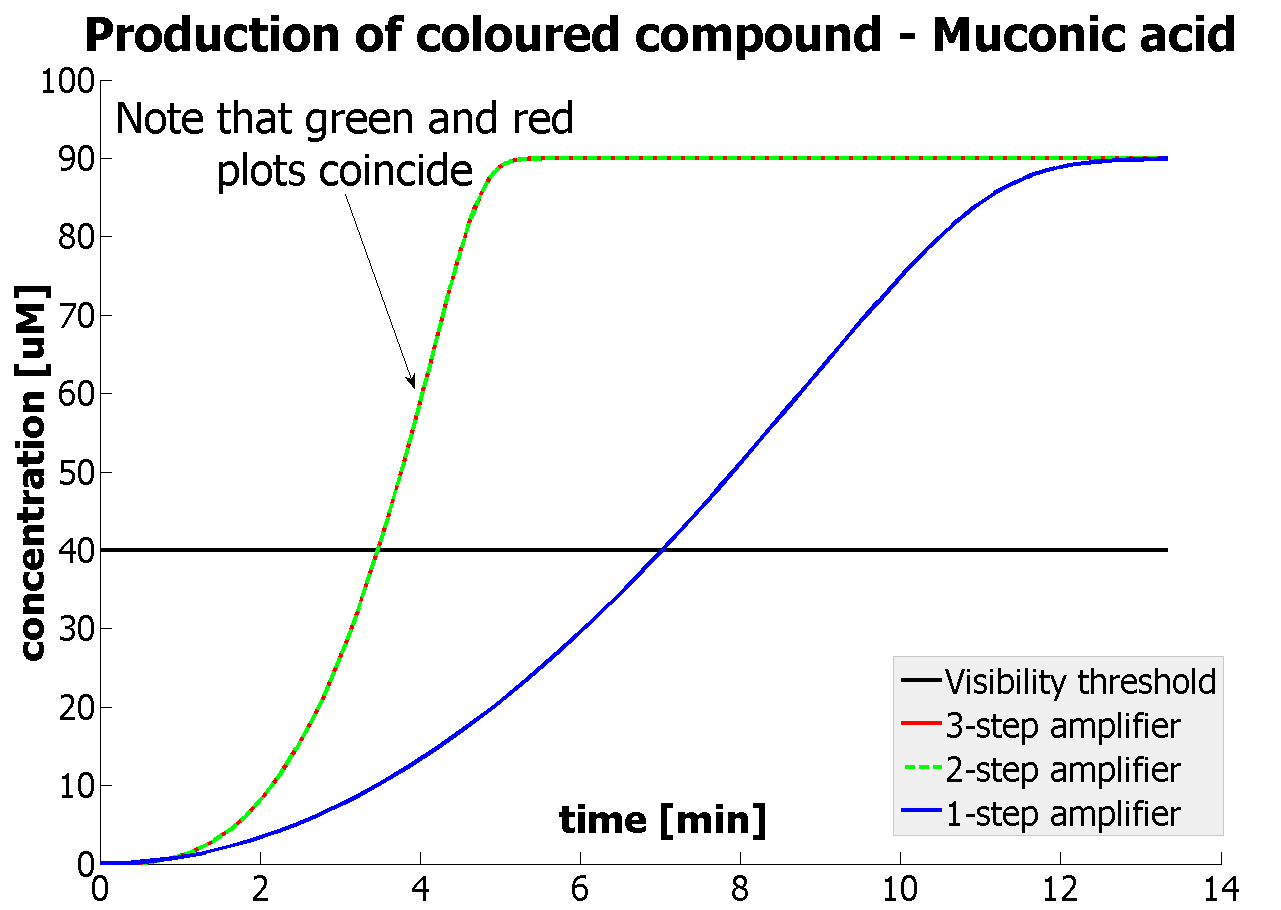
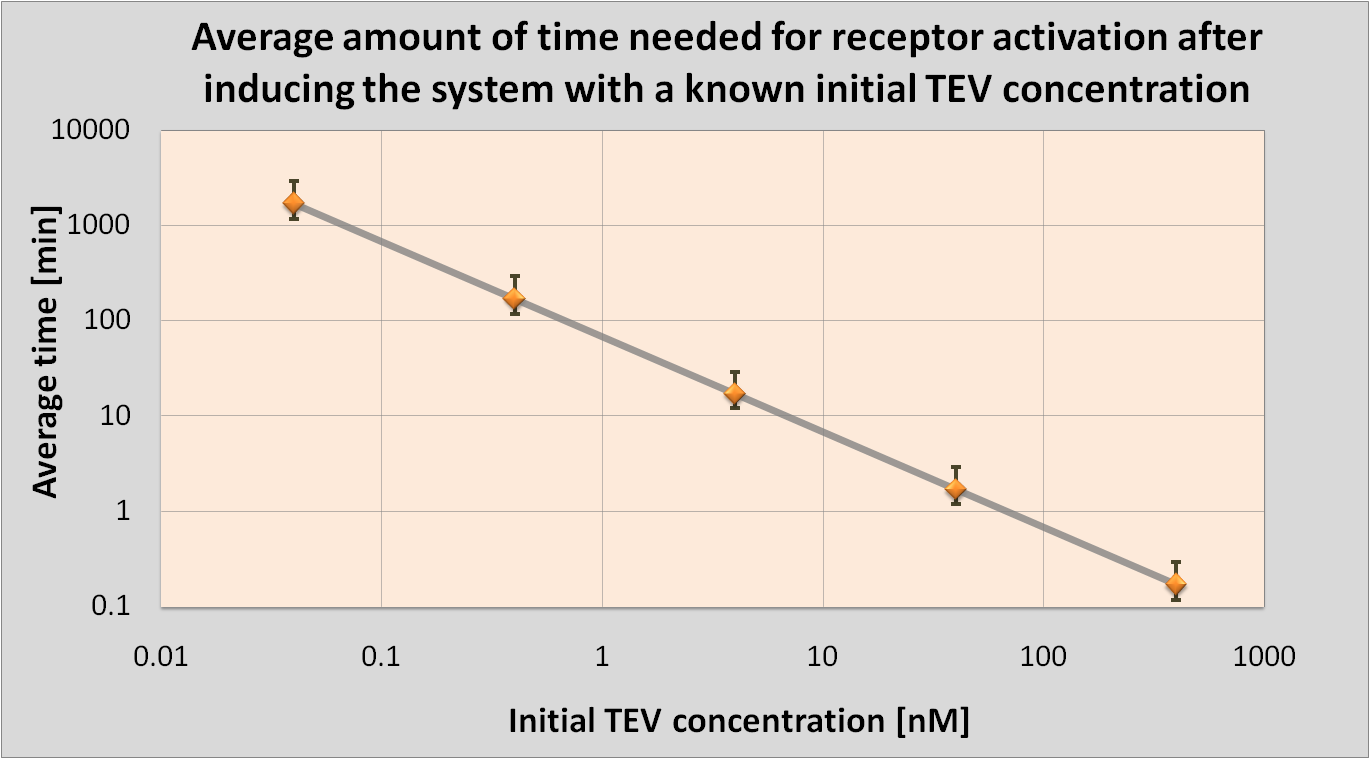
| A major part of the project consisted of modelling each module. This enabled us to decide which ideas we should implement. Look at the Fast Response page for a great example of how modelling has made a major impact on our design! | |
| Introduction to modelling |
In the process of designing our construct two major questions arose which could be answered by computer modelling:
|
| Results & Conclusions |
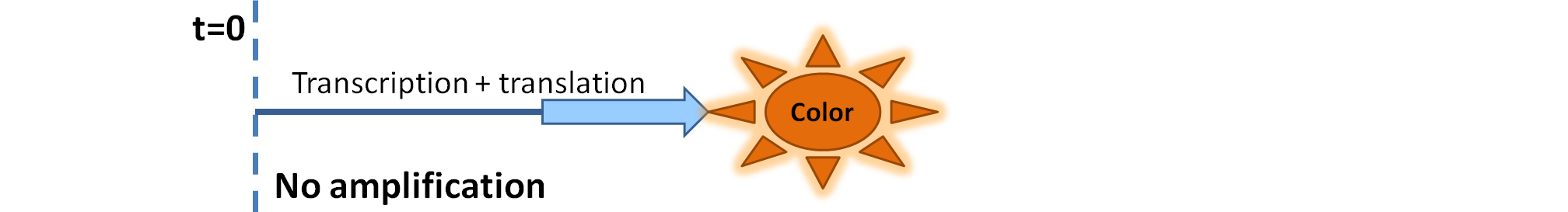
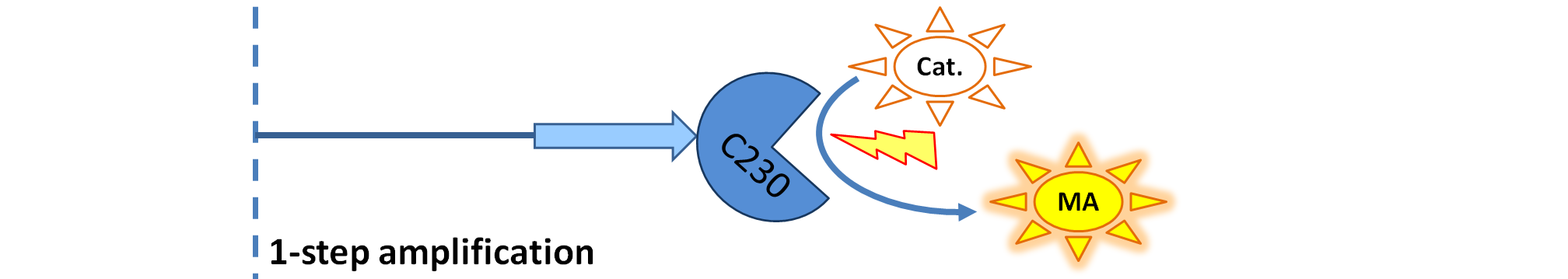
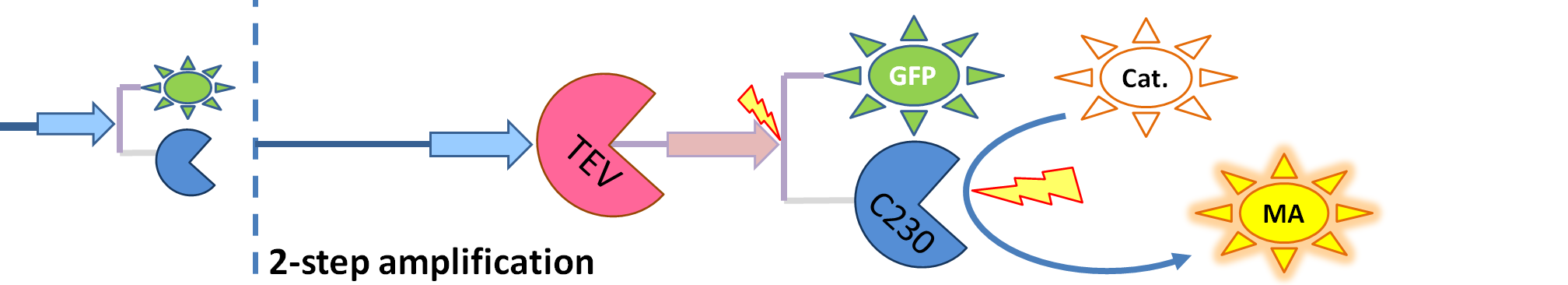
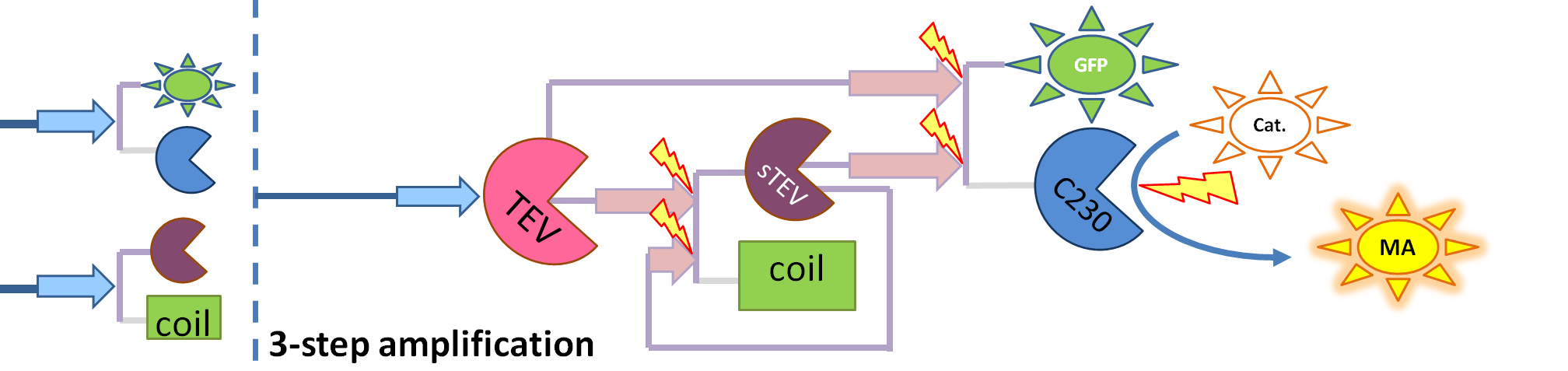
Output Amplification Model
Signalling Module Model Surface Protein Model
|
 "
"