Team:Harvard/flavor/notebook
From 2010.igem.org
(→07-06-2010 [ top ]) |
(→07-07-2010 [ top ]) |
||
| Line 698: | Line 698: | ||
==<html><a class="labnotebook" name="07-07-2010">07-07-2010</a></html> [ [[#top|top]] ]== | ==<html><a class="labnotebook" name="07-07-2010">07-07-2010</a></html> [ [[#top|top]] ]== | ||
| + | ===Sequencing=== | ||
| + | V0120 Plasmids containing the pENTCUP2 plant promoter, NOS terminator and NOS terminator + STOP were sent to GENEWIZ for sequencing. Sequencing results are expected tomorrow. | ||
| + | [[Image:sequencing_order.png|Sequencing Order]] | ||
| + | |||
| + | |||
| + | ===Cultures=== | ||
| + | 5 mL cultures were started from the ''YFP-2x'' construct from yesterday, as well as the B15 (StrepII) tag. | ||
| + | *2 x B21 E/X + Brazz E/S - C-Terminus | ||
| + | *2 x B21 S/P + Brazz X/P - N-Terminus | ||
| + | *2 x B21 E/X + Mira E/S - C-Terminus | ||
| + | *2 x B21 S/P + Mira X/P - N-Terminus | ||
| + | *2 x B15 Plate #1 | ||
| + | *2 x B15 Plate #2 | ||
| + | |||
| + | Cultures were placed in a 37°C incubator and left to shake overnight. | ||
| + | |||
| + | |||
| + | ===Primers for Wintergreen pathway parts=== | ||
| + | |||
| + | '''J45004''' | ||
| + | * Left Primer: 5' cctttctagaatggaagttgttgaagttcttca 3' | ||
| + | * Right Primer: 5' aaggctgcagcggccgctactagtttaatttattttggtcaagga 3' (last 5 bp omitted to meet 45 bp maximum) | ||
| + | |||
| + | '''J45017''' | ||
| + | * Left Primer: 5' cctttctagaatgaaaactcccgaagactgc 3' | ||
| + | * Right Primer: 5' aaggctgcagcggccgctactagtttattaggcgacgccgc 3' | ||
==<html><a class="labnotebook" name="07-08-2010">07-08-2010</a></html> [ [[#top|top]] ]== | ==<html><a class="labnotebook" name="07-08-2010">07-08-2010</a></html> [ [[#top|top]] ]== | ||
Revision as of 01:20, 25 October 2010

notebook calendar [ top ]
06-14-2010 [ top ]
- Miraculin and brazzein constructs due to arrive on Wednesday from Mr. Gene
BioBrick Transformation
BioBrick parts from the 2010 iGEM kit were transformed and grown in highly competent TURBO bacteria.
- Wintergreen Scent Pathway:
BBa_J45700 - entire pathway, Ampicillin
BBa_J45004 - BSMT1 only, Ampicillin
(Not in 2010 BB Kit: BBa_J45017 - PchB, PchA)
- Banana Scent Pathway:
BBa_J45250 - ATF3 + Promoter, Ampicillin?
BBa_J45014 - ATF3 only, Ampicillin
(Not in BB 2010 Kit: BBa_J45400 - BAT2 and THI3)
06-15-2010 [ top ]
Primer Designs for Agrobacterium Vector
- Primers were designed to amplify from the Expression Series pORE Agrobacterium plasmid (e3) 1)the pENTcup2 promoter, 2) the tNOS stop sequence, 3) the tNOS stop sequence + additional stop codon and 4) the pHLP promoter.
Note: the NOSterm_BB_R primer works for both tNOS and tNOS+STOP codon sequences.
pENTcup2_BB_F CCTTTCTAGAGGGATCTTCTGCAAGCATCT
pENTcup2_BB_R AAGGCTGCAGCGGCCGCTACTAGTTCCGGTGGGTTTTGAGGT
STOP_NOSterm_BB_F CCTTTCTAGATGAGATCGTTCAAACATTTGG
NOSterm_BB_F CCTTTCTAGAGATCGTTCAAACATTTGGCA
NOSterm_BB_R AAGGCTGCAGCGGCCGCTACTAGTGATCTAGTAACATAGATGACA
pHPL_BB_F CCTTTCTAGAAACGTGGATACTTGGCAGTG
pHPL_BB_R AAGGCTGCAGCGGCCGCTACTAGTCTTTTGAGCTTAGAGGTTTTT
BioBrick Transformation
- Colonies were observed after overnight culture growth, but in low quantities.
- J45700 and J45004 (both Wintergreen Pathway) showed minimal number of colonies on both 10μL and 100μL cultures.
- J45250 and J45014 (both Banana Pathway) showed no colonies on either the 10μL or 100μL cultures.
Codon Usage in Arabidopsis
Using http://gcua.schoedl.de/
Valencene Extraction
Used RNeasy Plant Mini Kit to extract RNA from the flavedo of an Organic Valencia Orange.
06-16-2010 [ top ]
MiniPrep
- MiniPrep of Wintergreen parts from Registry (J45004 and J45700) following Qiagen MiniPrep protocol. MiniPrep three samples of each part.
- DNA concentrations:
J45004-1: 59.1 ng/μL
J45004-2: 72.9 ng/μL
J45004-3: 88.1 ng/μL
J45700-1: 146.7 ng/μL
J45700-2: 187.4 ng/μL
J45700-3: 175.5 ng/μL
Digestion with Enzymes
- For J45004, we added: 9μL DNA, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast).
- For J45700, we added 5μL DNA, 4μL H2O, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast).
- We let mixtures sit for 30 minutes due to the use of xbaI restriction enzyme (slow). After the 30 minutes, we added 2.5μL dye to each mix.
Gel
- We loaded 12.5μL of 1kb ladder to well 1 of the gel (numbered left to right). We then loaded 12.5μL of J45004-1, J45004-2, and J45004-3 to wells 2,3, and 4, respectively. We loaded J45700-1, J45700-2, J4500-3 in wells 5, 6, and 7, respectively (see images below for well locations).
- Ran on 1% agarose gel.
06-17-2010 [ top ]
PCR Purification of pORE Vector Parts
Following QIAgen PCR Purification Kit Protocol the following PCR products were purified:
- pENTCUP2 Promoter - 16.1 ng/μL
- NOSterminator Sequence - 76.3 ng/μL
- STOP codon + NOSterminator Sequence - 76.5 ng/μL
To do: follow up with restriction digest ✓ and Agarose gel ✓ to confirm PCR and PCR purification.
Restriction Digest and Agarose Gel
Restriction Digest reactions were set up as follows:
pENTCUP2 Promoter:
- 27μL PCR product
- 3μL Loading Buffer w/ dye
- 1μL Xba1 Fast Enzyme
- 1μL Pst1 Fast Enzyme
NOSterm and NOSterm + STOP:
- 9μL PCR product
- 1μL Loading Buffer w/ dye
- 0.5μL Xba1 Fast Enzyme
- 0.5μL Pst1 Fast Enzyme
BioBrick Plasmid V0120 (Ampicillin Resistance):
- 3μL DNA
- 1μL Loading Buffer w/ dye
- 6μL diH2O
- 0.5μL Xba1 Fast Enzyme
- 0.5μL Pst1 Fast Enzyme
Reactions were allowed to proceed for 45 minutes at 37°C.
06-18-2010 [ top ]
06-21-2010 [ top ]
06-22-2010 [ top ]
06-23-2010 [ top ]
06-24-2010 [ top ]
06-25-2010 [ top ]
We began this week trying to ligate the pENTCUP2 promoter, NosT terminator, and NosT terminator plus stop codon into the V0120 vector. We ran into several problems during this process.
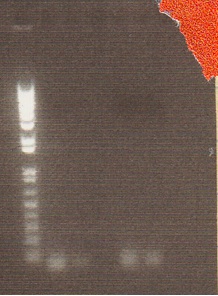
We had previously digested the three inserts as well as the promoter with XbaI and PstI fast digest enzymes, ligated, and transformed the ligated vectors into e. coli. Unfortunately, we did not find any colonies of cells after over 14 hours of incubation. Furthermore, on Friday June 18th, after digesting all three inserts and vector and running them on a gel, we did not see any NosT + stop DNA. Therefore, on Monday June 21, we began by digesting more NosT + stop and V0120 vector with xba1 and pstI fast digestive enzymes. The gel showed DNA length to be consistent with the given parts. (See gel below)
- Gel Lanes (both inserts and vector were digested with xbaI/pstI): 1. 1kb plus ladder. 2. pENTCUP2 promoter. 3. NosT terminator. 4. NosT + stop terminator 5. V0120 digested.
- Gel Lanes (both inserts and vector were digested with xbaI/pstI): 1. 1kb plus ladder. 2. NosT + stop terminator. 3. V0120 digested.
Upon gel extraction and gel purification, we ligated the inserts into the vectors with a 2:1 insert to vector ratio (previously we had been using a 4:1 insert to vector ratio). We then transformed the plasmids into e.coli and plated; no colony growth was seen the next morning.
We used Team Vector's R3 ligation (they have successfully ligated and transformed) as a positive control. However, the bacteria containing the ligated plasmids were plated on Ampicillin plates, which turned out to be the wrong resistance.
Troubleshooting
In order to get a better understanding of the problem with our ligation, we ran a diagnostic gel with the V0120 backbone and vectors B15 and B21.
Gel Lanes: 1. 1 kb plus ladder 2. v0120 undigested 3. v0120 eco/spe 4. v0120 eco/xba 5. v0120 spe/pstI 6. v0120 xba/pst 7. v0120 ecoRI 8. v0120 xba1 9. v0120 pstI 10. v0120 spe 11. blank 12. B15 (StrepII tag) xba/pstI 13. B21 (YFP) xba/pstI
We concluded from the gel that xba1 is the problem in our ligation step. The pattern of bands in the v0120 xba1/pstI digest along with the v0120 xba1 digest illustrate that xba1 is not cutting correctly.
We began a slow enzyme xba1/pstI digest on Friday, but the gel was run in mismatching buffers (TAE and TBE).
- Gel Lanes (inserts and vector digested with xba1/pstI): 1. 1 kb plus ladder. 2. pENTCUP2 3. NosT 4. NosT & stop 5. v0120
Valencene
Digestion of PCR products aimed to isolate valencene gene showed very short length on gel. These are not the correct size and are most likely primer dimers. PCR products were digested with EcoRI/SpeI. 5 μL of each sample was digested with pstI to be sure that the pstI site was in the sequence of DNA.
- Gel Lanes: 1. 1 kb plus ladder. 2. Flavedo EcoRI/SpeI. 3. Flavedo PstI 4. Open 5. Fruit EcoRI/SpeI. 6. Fruit pstI
06-28-2010 [ top ]
Miniprep of Miraculin and Brazzein Constructs
- Protocol used was from the Qiagen Miniprep Kit
| Name | Concentration (ng/μL) |
|---|---|
| Miraculin 1 | 159.6 |
| Miraculin 2 | 182.2 |
| Brazzein 1 | 91.2 |
| Brazzein 2 | 33.7 |
DTL (Digestion, Ligation, Transformation) of The Big Three (pENTCUP2, NOSt, NOSt + STOP)
| pENTCUP2 | NOSt | NOSt+STOP | B15 | |
|---|---|---|---|---|
| DNA | 4 | 7 | 12 | 7 |
| NEB Buffer 3 (10x) | 2 | 2 | 2 | 2 |
| diH2O | 10 | 7 | 2 | 7 |
| Xba1 | 1 | 1 | 1 | 1 |
| Pst1 | 1 | 1 | 1 | 1 |
| BSA (10x) | 2 | 2 | 2 | 2 |
- Digestions were left for 1:30 at 37°C
- 2.2 μL of DNA Loading Buffer were added to each reaction and loaded onto a 1% Agarose gel (TAE buffer). Gel was ran at 125 V for 30 min.
| Lane | Contents |
|---|---|
| Lane 1 | 1KB Plus Ladder |
| Lane 2 | pENTCUP2 |
| Lane 3 | NOSt |
| Lane 4 | NOSt+STOP |
| Lane 5 | B15 digested |
| Lane 6 | B15 undigested |
- The undigested B15 in lane 6 appears as two bands on account of supercoiling of the B15 plasmid.
- Bands in lanes 2 - 5 were cut out and purified using a Qiagen Gel Purification kit
- The gel purification was done with 300 μL Buffer PB and 400 μL Buffer QG
| Name | Concentration (ng/μL) |
|---|---|
| pENTCUP2 | 16.5 |
| NOSt | 10.2 |
| NOSt+STOP | 0.4 |
| B15 (backbone vector) | 10.9 |
- Note the very low concentration of NOSt+STOP. It is uncertain as to what caused such a concentration, but transformation proceeded anyways with max volume
- The ligation reactions were preformed at at 2:1 (insert to backbone) Molar Ratio
- The sole exception being the NOSt+STOP ligation, in which the lack of product forced us to use 1.8 ng DNA
| pENTCUP2 | NOSt | NOSt+STOP | Control | |
|---|---|---|---|---|
| Insert | 1 | 1 | 1 | 1 |
| Vector | 2 | 2 | 2 | 2 |
| Buffer (10x) | 5 | 5 | 5 | 5 |
| Ligase | 1 | 1 | 1 | 1 |
| H2O | 11 | 11 | 0 | 12 |
- Ligation reactions were preformed as per the Silver Lab ligation protocol (with the differences in concentration and ratio made as per the table above).
- Transformations to E. Coli were preformed as per the Silver lab transformation protocol with ampicillin plates.
- Plates were left overnight.
06-29-2010 [ top ]
Results of ligation and transformation of The Big Three (pENTCUP2, NosT, NosT + stop) with B15 backbone into E.coli
A few colonies were found on the pENTCUP2 + Backbone plate, NosT + stop and backbone plate, and b15 backbone control plate. Colonies were picked from the plates and placed in 3ml of LB and agarose in a culture tube. The tubes were placed in the 37°C shaker for 8 hours. 2 mL of LB+AMP was added to the cultures, bring the total volume to 5 mL of LB+AMP. Cultures were left to shake overnight.
DLT of the Sweethearts - Miraculin and Brazzein (and B21)
| Miraculin | Brazzein | B21 | |
|---|---|---|---|
| DNA | 6 | 11 | 7 |
| FD Buffer (10x) | 2 | 2 | 2 |
| diH2O | 10 | 5 | 9 |
| EcoRI | 1 | 1 | 1 |
| SpeI | 1 | 1 | 1 |
Placed in 37°C waterbath for 20 minutes.
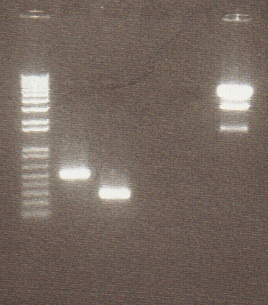
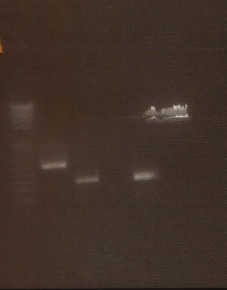
Digestion Gel
Gel Lanes:
1. 1 kb plus ladder
2. Miraculin digested with EcoRI/SpeI
3. Brazzein digested with EcoRI/SpeI
4. B21 digested with EcoRI/SpeI
5. 1 kb plus ladder
| Name | Concentration (ng/μL) |
|---|---|
| Miraculin | 9.5 |
| Brazzein | 10.2 |
| B21 Vector (V0120) | 8.4 |
| Miraculin | Brazzein | Control | |
|---|---|---|---|
| Insert | 3.5 | 0.75 | 0 |
| Vector | 6 | 6 | 6 |
| Buffer (10x) | 2 | 2 | 2 |
| Ligase | 1 | 1 | 1 |
| H2O | 6.5 | 10.25 | 0 |
- The ligation of Miraculin was done at a 3:1 ratio (insert to vector) due to its larger size (~700 bp)
- The ligation of Brazzein was done at a 2:1 ratio due to its smaller size (~200 bp)
06-30-2010 [ top ]
Miraculin, Brazzein
- Colonies did grow!
- Started 5 mL cultures of Miraculin and Brazzein biobrick constructs (5 cultures each)
The Big Three
- Began miniprep of cultures of pENT, NOSt+STOP and our Control (for experimental purposes)
| OD | |
|---|---|
| pENTCUP2 #1 | 248.5 ng/μL |
| pENTCUP2 #2 | 447.7 ng/μL |
| NosT + stop #1 | 427.7 ng/μL |
| NosT + stop #2 | 303.9 ng/μL |
| Control #1 | 366.8 ng/μL |
| Control #2 | 359.2 ng/μL |
Digestion
- We digested miniprep samples with xbaI/pstI fast digest enzymes in order to determine if ligation was successful.
| Control #1 | Control #2 | pENTCUP2 #1 | pENTCUP2 #2 | NosT & stop #1 | NosT & stop #2 | |
|---|---|---|---|---|---|---|
| DNA | 2 | 2 | 3 | 2 | 2 | 3 |
| Green FD buffer | 2 | 2 | 2 | 2 | 2 | 2 |
| diH2O | 14 | 14 | 13 | 14 | 14 | 13 |
| xbaI | 1 | 1 | 1 | 1 | 1 | 1 |
| pstI | 1 | 1 | 1 | 1 | 1 | 1 |
Digestion mixes were placed in 37°C water bath for approximately 1 hr.
Big Three PCR digestion
- We simultaneously digested PCR purified pENTCUP2, NosT, and NosT & stop inserts with xbaI/pstI. This way if the above digestion showed the ligation of the inserts into v0120 backbone was not successful, we can continue with these inserts.
| pENTCUP2 | NosT | NosT & stop | |
|---|---|---|---|
| DNA | 4 | 7 | 2 |
| Green FD buffer | 2 | 2 | 2 |
| diH2O | 12 | 9 | 14 |
| xbaI | 1 | 1 | 1 |
| pstI | 1 | 1 | 1 |
Placed in 37°C water bath for 20 minutes.
07-01-2010 [ top ]
- We made glycerol stocks from our Miraculin & v0120 and Brazzein & v0120. 333 μL cultures with 666μL glycerol and placed in -80°C freezer.
- We miniprepped our Miraculin and v0120 cultures and our Brazzein and v0120 cultures. Miniprep done per Qiagen protocol.
ODs
| Sample | OD |
|---|---|
| Miraculin and v0120 #1 | 171.3ng/μL |
| Miraculin and v0120 #2 | 202.0ng/μL |
| Miraculin and v0120 #3 | 106.4ng/μL |
| Miraculin and v0120 #4 | 115.6ng/μL |
| Miraculin and v0120 #5 | 40.5ng/μL |
| Brazzein and v0120 #1 | 41.7ng/μL |
| Brazzein and v0120 #2 | 42.8ng/μL |
| Brazzein and v0120 #3 | 38.8ng/μL |
| Brazzein and v0120 #4 | 0ng/μL |
| Brazzein and v0120 #5 | N/A |
Valencia orange surgery (part two)
- RNA extraction per Qiagen RNAeasy protocol.
- We made sure to apply RNAzap (RNAase-free) spray to all surfaces and instruments.
ODs
| Sample | OD |
|---|---|
| RNA Flavedo extraction #1 | 93.1ng/μL |
| RNA Flavedo extraction #2 | 28.7ng/μL |
07-02-2010 [ top ]
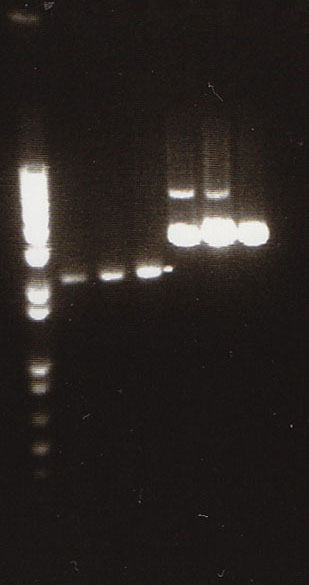
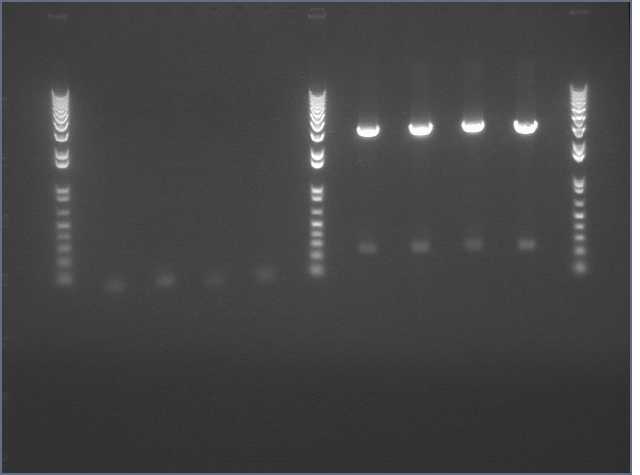
NosT + stop and v0120
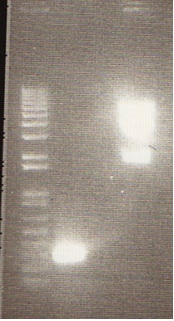
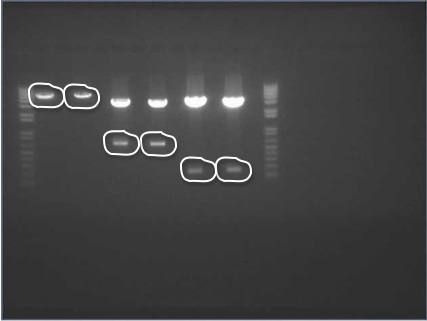
We ran an xba/pstI fast enzyme digest on our miniprepped NosT + stop and v0120 plasmid. The gel was consistent with a successful ligation. (See lanes 7-10 in gel image below).
Valencene
After running a reverse transcriptase reaction on our RNA extraction, we ran PCR on the cDNA with Valencene specific primers. The gel did not show the proper fragment length of the Valencene gene. Instead, it appears primer dimers formed again. (See lanes 2-5)
07-05-2010 [ top ]
07-06-2010 [ top ]
Tagging Miraculin and Brazzein with YFP and StrepII tags
Tag: -E--N--X--STREP/YFP--S--N--P-
Insert: E--N--X--Miraculin/Brazzein--S--N--P-
- The 'tag' biobrick (either YFP or STREPII) was used as the vector; Miraculin and Brazzein were used as the insert.
- Tags were ligated to both the N- and C-terminal of Miraculin and Brazzein
| Vector | Insert | |
|---|---|---|
| N-Terminus | Spe1/Pst1 | Xba1/Pst1 |
| C-Terminus | EcoR1/Xba1 | EcoR1/Spe1 |
| B21 S/P | B21 E/X | Miraculin X/P | Miraculin E/S | Brazzein X/P | Brazzein E/S | |
|---|---|---|---|---|---|---|
| diH2O | 13 | 13 | 11 | 11 | 0 | 0 |
| Green FD buffer (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| DNA | 3 | 3 | 5 | 5 | 16 | 16 |
| Spe1 | 1 | - | - | 1 | - | 1 |
| PstI | 1 | - | 1 | - | 1 | - |
| EcoR1 | - | 1 | - | 1 | - | 1 |
| Xba1 | - | 1 | 1 | - | 1 | - |
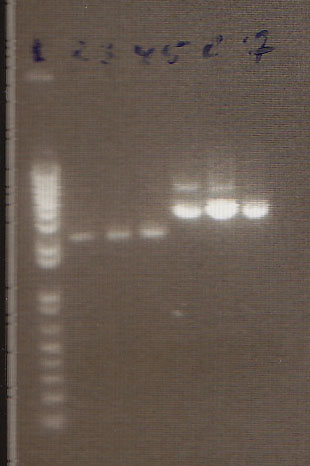
Gel
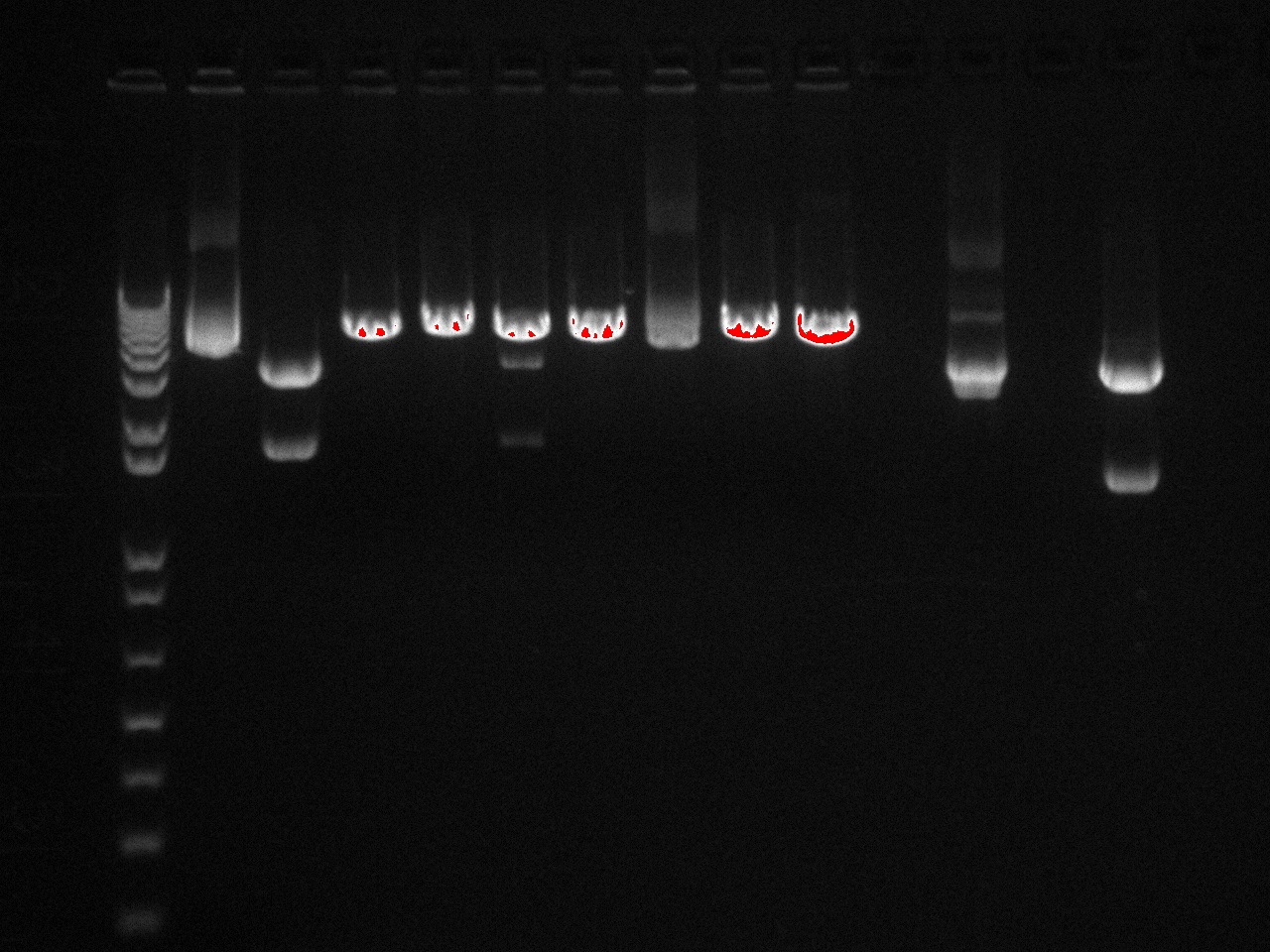
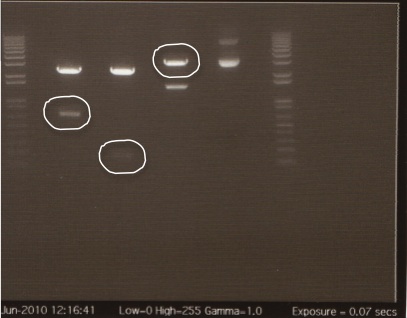
The gel showed DNA fragments consistent with Miraculin and Brazzein. The digestion of B21 appeared to be succesful, but the DNA sequence cut out was too small to see on the gel.
Gel Lanes:
1. 1 kb plus ladder
2. B21 speI/pstI
4. B21 ecoRI/xbaI
6. Miraculin xbaI/pstI
8. Miraculin ecoRI/speI
10. Brazzein xbaI/pstI
12. Brazzein ecoRI/pstI
14. 1kb plus ladder
We extracted the circled bands and gel purified the gel per the Qiagen gel purification protocol.
ODs of gel purified DNA
B21 speI/pstI: 15.9 ng/μL
B21 ecoRI/xbaI: 11.0 ng/μL
Miraculin xbaI/pstI: 9.7 ng/μL
Miraculin ecoRI/speI: 7.1 ng/μL
Brazzein xbaI/pstI: 2.6 ng/μL
Brazzein ecoRI/pstI: 5.0 ng/μL
Ligation
We did 6 different ligation reactions. The chart below shows the different reactions. Ligation reactions were left at room temperature for 15 minutes.
| Miraculin xbaI/pstI w/ B21 speI/pstI | Miraculin ecoRI/speI w/ B21 ecoRI/xbaI | Brazzein xbaI/pstI w/ B21 speI/pstI | Brazzein ecoRI/speI w/ B21 ecoRI/xbaI | Control B21 speI/pstI | Control B21 ecoRI/xbaI | |
|---|---|---|---|---|---|---|
| DNA Insert | 2 | 3 | 3 | 1 | 0 | 0 |
| T4 DNA ligase buffer (10x) | 2 | 2 | 2 | 2 | 2 | 2 |
| diH2O | 12 | 9 | 11 | 11 | 14 | 12 |
| T4 DNA ligase | 1 | 1 | 1 | 1 | 1 | 1 |
| DNA Backbone | 1 | 1 | 1 | 1 | 1 | 1 |
Transformation
We mixed 5μL ligation mix with 15μL Turbo e. coli cells. These were placed in ice for 30 minutes and then heat shocked in 42°C water bath for 30 seconds. The cells were placed back on ice for 2 minutes. 170 μL of SOC broth was added to each Eppendorf tube. The transformed e. coli were then plated on LB Ampicillin plates and left in the 37°C incubator overnight.
07-07-2010 [ top ]
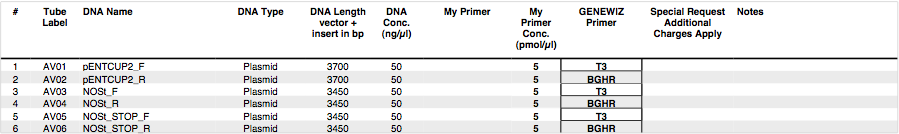
Sequencing
V0120 Plasmids containing the pENTCUP2 plant promoter, NOS terminator and NOS terminator + STOP were sent to GENEWIZ for sequencing. Sequencing results are expected tomorrow.
Cultures
5 mL cultures were started from the YFP-2x construct from yesterday, as well as the B15 (StrepII) tag.
- 2 x B21 E/X + Brazz E/S - C-Terminus
- 2 x B21 S/P + Brazz X/P - N-Terminus
- 2 x B21 E/X + Mira E/S - C-Terminus
- 2 x B21 S/P + Mira X/P - N-Terminus
- 2 x B15 Plate #1
- 2 x B15 Plate #2
Cultures were placed in a 37°C incubator and left to shake overnight.
Primers for Wintergreen pathway parts
J45004
- Left Primer: 5' cctttctagaatggaagttgttgaagttcttca 3'
- Right Primer: 5' aaggctgcagcggccgctactagtttaatttattttggtcaagga 3' (last 5 bp omitted to meet 45 bp maximum)
J45017
- Left Primer: 5' cctttctagaatgaaaactcccgaagactgc 3'
- Right Primer: 5' aaggctgcagcggccgctactagtttattaggcgacgccgc 3'
 "
"