Team:Harvard/flavor/notebook
From 2010.igem.org
(Difference between revisions)
(→06-16-2010 [ top ]) |
|||
| Line 81: | Line 81: | ||
==<html><a class="labnotebook" name="06-16-2010">06-16-2010</a></html> [ [[#top|top]] ]== | ==<html><a class="labnotebook" name="06-16-2010">06-16-2010</a></html> [ [[#top|top]] ]== | ||
| + | ===MiniPrep=== | ||
| + | * MiniPrep of Wintergreen parts from Registry (J45004 and J45700) following Qiagen MiniPrep protocol. MiniPrep three samples of each part. | ||
| + | ** DNA concentrations: | ||
| + | J45004-1: 59.1 ng/μL | ||
| + | J45004-2: 72.9 ng/μL | ||
| + | J45004-3: 88.1 ng/μL | ||
| + | |||
| + | J45700-1: 146.7 ng/μL | ||
| + | J45700-2: 187.4 ng/μL | ||
| + | J45700-3: 175.5 ng/μL | ||
| + | ===Digestion with Enzymes=== | ||
| + | * For J45004, we added: 9μL DNA, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast). | ||
| + | * For J45700, we added 5μL DNA, 4μL H2O, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast). | ||
| + | * We let mixtures sit for 30 minutes due to the use of xbaI restriction enzyme (slow). After the 30 minutes, we added 2.5μL dye to each mix. | ||
| + | |||
| + | ===Gel=== | ||
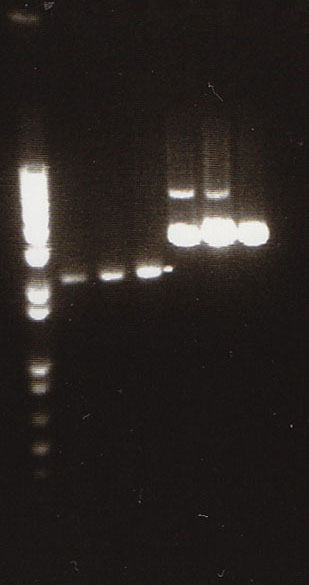
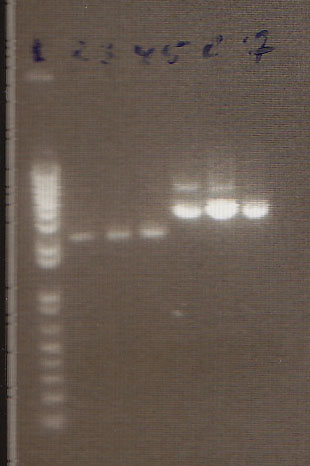
| + | * We loaded 12.5μL of 1kb ladder to well 1 of the gel (numbered left to right). We then loaded 12.5μL of J45004-1, J45004-2, and J45004-3 to wells 2,3, and 4, respectively. We loaded J45700-1, J45700-2, J4500-3 in wells 5, 6, and 7, respectively (see images below for well locations). | ||
| + | * Ran on 1% agarose gel. | ||
| + | |||
| + | [[Image:J45004_J45700gel1.jpg|150px|BBa_J45700]] | ||
| + | [[Image:J45004_J45700gel2.jpg|150px|BBa_J45700]] | ||
==<html><a class="labnotebook" name="06-17-2010">06-17-2010</a></html> [ [[#top|top]] ]== | ==<html><a class="labnotebook" name="06-17-2010">06-17-2010</a></html> [ [[#top|top]] ]== | ||
Revision as of 00:27, 25 October 2010

notebook calendar [ top ]
06-14-2010 [ top ]
- Miraculin and brazzein constructs due to arrive on Wednesday from Mr. Gene
BioBrick Transformation
BioBrick parts from the 2010 iGEM kit were transformed and grown in highly competent TURBO bacteria.
- Wintergreen Scent Pathway:
BBa_J45700 - entire pathway, Ampicillin
BBa_J45004 - BSMT1 only, Ampicillin
(Not in 2010 BB Kit: BBa_J45017 - PchB, PchA)
- Banana Scent Pathway:
BBa_J45250 - ATF3 + Promoter, Ampicillin?
BBa_J45014 - ATF3 only, Ampicillin
(Not in BB 2010 Kit: BBa_J45400 - BAT2 and THI3)
06-15-2010 [ top ]
Primer Designs for Agrobacterium Vector
- Primers were designed to amplify from the Expression Series pORE Agrobacterium plasmid (e3) 1)the pENTcup2 promoter, 2) the tNOS stop sequence, 3) the tNOS stop sequence + additional stop codon and 4) the pHLP promoter.
Note: the NOSterm_BB_R primer works for both tNOS and tNOS+STOP codon sequences.
pENTcup2_BB_F CCTTTCTAGAGGGATCTTCTGCAAGCATCT
pENTcup2_BB_R AAGGCTGCAGCGGCCGCTACTAGTTCCGGTGGGTTTTGAGGT
STOP_NOSterm_BB_F CCTTTCTAGATGAGATCGTTCAAACATTTGG
NOSterm_BB_F CCTTTCTAGAGATCGTTCAAACATTTGGCA
NOSterm_BB_R AAGGCTGCAGCGGCCGCTACTAGTGATCTAGTAACATAGATGACA
pHPL_BB_F CCTTTCTAGAAACGTGGATACTTGGCAGTG
pHPL_BB_R AAGGCTGCAGCGGCCGCTACTAGTCTTTTGAGCTTAGAGGTTTTT
BioBrick Transformation
- Colonies were observed after overnight culture growth, but in low quantities.
- J45700 and J45004 (both Wintergreen Pathway) showed minimal number of colonies on both 10μL and 100μL cultures.
- J45250 and J45014 (both Banana Pathway) showed no colonies on either the 10μL or 100μL cultures.
Codon Usage in Arabidopsis
Using http://gcua.schoedl.de/
Valencene Extraction
Used RNeasy Plant Mini Kit to extract RNA from the flavedo of an Organic Valencia Orange.
06-16-2010 [ top ]
MiniPrep
- MiniPrep of Wintergreen parts from Registry (J45004 and J45700) following Qiagen MiniPrep protocol. MiniPrep three samples of each part.
- DNA concentrations:
J45004-1: 59.1 ng/μL
J45004-2: 72.9 ng/μL
J45004-3: 88.1 ng/μL
J45700-1: 146.7 ng/μL
J45700-2: 187.4 ng/μL
J45700-3: 175.5 ng/μL
Digestion with Enzymes
- For J45004, we added: 9μL DNA, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast).
- For J45700, we added 5μL DNA, 4μL H2O, 1μL buffer, 1μL xbaI restriction enzyme (slow), and .5μL pstI restriction enzyme (fast).
- We let mixtures sit for 30 minutes due to the use of xbaI restriction enzyme (slow). After the 30 minutes, we added 2.5μL dye to each mix.
Gel
- We loaded 12.5μL of 1kb ladder to well 1 of the gel (numbered left to right). We then loaded 12.5μL of J45004-1, J45004-2, and J45004-3 to wells 2,3, and 4, respectively. We loaded J45700-1, J45700-2, J4500-3 in wells 5, 6, and 7, respectively (see images below for well locations).
- Ran on 1% agarose gel.
 "
"