Team:Imperial College London/Lab Diaries/XylE team
From 2010.igem.org
(→Week 7) |
|||
| Line 102: | Line 102: | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| Week 6 |
|- | |- | ||
| | | | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
| - | <table width=" | + | <table width="850px" border="0"> |
<tr> | <tr> | ||
<td style="background-color:#FFFF66"> | <td style="background-color:#FFFF66"> | ||
| Line 202: | Line 178: | ||
</html> | </html> | ||
| + | |||
| + | '''Thursday, 12-Aug-2010''' | ||
| + | * Anealing DNA strands of J23101 promoter in a waterbath | ||
| + | we constructed the standard E.coli promoter J23101 with sticky ends. These ends are complementary to restriction sites made by EcoRI and SpeI enzyme. This promoter will be later used in 3A assemply to construct a promoter-RBS-XylE design in a psB1C3 vector. E.coli will be transformed with this final construct plasmid to assess XylE activity and characterization. It will also be one of the submitted biobricks. | ||
| + | |||
| + | * Prepared two overnight cultures of XylE transormed E.coli (one 50microliters and one of 450 microliters) | ||
| + | these cultures are going to be used tomorrow for mini-prepping. Miniprep will allow us to isolate E.coli's plasmid DNA(which contains the XylE gene). | ||
| + | |||
| + | '''Friday, 13-Aug-2010''' | ||
| + | * Mini-prep of XylE transformed E.coli | ||
| + | Mini-prep is usually used to confirm that our gene of interest has not been changed in any way, as the isolated plasnid id sent for sequencing. However, since XylE was taken from the registry, we assume that it is fine and no sequencing is required. The mini-prep will later be used for the midi-prep (that gives out higher yeilds of DNA needed for cloning). | ||
| + | * Gel analysis of plasmid DNA retreived from mini-prep of XylE transformed E.coli, cut with restriction enzymes. From light to the left, 50micrograms digested DNA : 50 undigested DNA : 450 digested DNA : 450 undigested DNA. In lanes 1 and 3 the smaller band has a size of about 1kB which corresponds to RBS-XylE gene. The bigger bands are the cut vectors. In lanes 2 and 4 is the uncut biobrick from the registry. It appears smaller on the gel than it actually is as circular DNA travels faster through the pores of agarose gel rather than linearised DNA. | ||
| + | |} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| Week 7 | ||
| + | |- | ||
| + | | | ||
| + | [[Image:IC_10-08-13_Digest_image.jpg|thumb|center|400px]] | ||
| + | |||
| + | <html> | ||
| + | <table width="850px" border="0"> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFFF66"> | ||
| + | <b>Week 7</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Monday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Tuesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Wednesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Thursday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Friday</b> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Morning</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>midi-prep XylE E.coli (2hrs)</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>gel extraction kit on XylE gene trapped in agarose </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>3A assemply: make replica plates (overnight)</li> | ||
| + | <li>Catechol assay of E.coli </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Mini-prep XylE E.coli</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Midi-prep XylE E.coli</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:50px;text-align:top;"><b>Lunch Break</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>restriction digestion of XylE for 3A assemply</li> | ||
| + | <li>gel purification of XylE from rstriction digestion</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>3A assemply of vector, XylE and J23101 promoter </li> | ||
| + | <li>above construct transformed in E.coli</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>*primers arrived!</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>PCR extension of XylE and GFP, round 1</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| - | + | '''Monday, 16-Aug-2010''' | |
| - | * | + | * Midi-prepped the XylE-transformed E.coli. The DNA yeild from the midi-prep was 134micrograms as determined by spectrophotometry. This is the XylE that is going to be used for all further experiments. |
| - | * | + | * Restriction digestion of midi-prepped XylE by Xbal and PstI to prepare it for 3A assemply. (with J23101 promoter and PSB1C3 vector) |
| - | * | + | * Gel analysis of the restriction digestion mixture to isolate XylE gene |
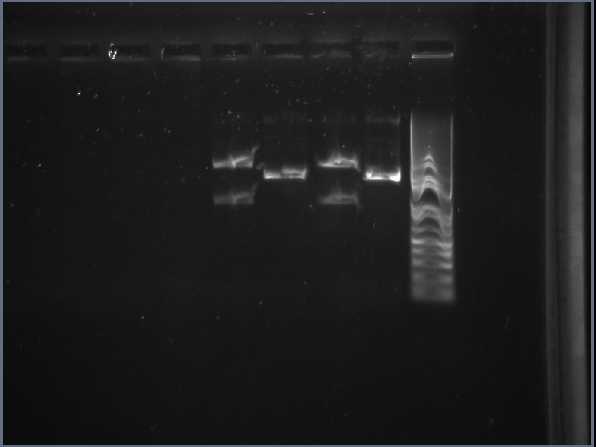
Midi-prep XylE digestion with xbaI and PstI The smaller size bands at lanes 2 & 3 are the ones that are going to be cut out and used in gel purification to extract the XylE gene (with sticky ends for XbaL and PstI). | Midi-prep XylE digestion with xbaI and PstI The smaller size bands at lanes 2 & 3 are the ones that are going to be cut out and used in gel purification to extract the XylE gene (with sticky ends for XbaL and PstI). | ||
| - | + | * I made an overnight culture of Bacillus | |
[[Image:IC_Midi-prep_XylE_digestion_with_xbaI_and_PstI.jpg|thumb|center|400px|Midi-prep XylE digestion with xbaI and PstI The smaller size bands at lanes 2 & 3 are the ones that are going to be cut out and used in gel purification to extract the XylE gene (with sticky ends for XbaL and PstI).]] | [[Image:IC_Midi-prep_XylE_digestion_with_xbaI_and_PstI.jpg|thumb|center|400px|Midi-prep XylE digestion with xbaI and PstI The smaller size bands at lanes 2 & 3 are the ones that are going to be cut out and used in gel purification to extract the XylE gene (with sticky ends for XbaL and PstI).]] | ||
| - | + | '''Tuesday, 17-Aug-2010''' | |
| - | * | + | * Uusing the Gel Extraction Kit, we isolated the restriction enzyme cut XylE gene from the agarose gel lamp. |
| - | * | + | * Gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine their ratios for 3A assemply ligation |
| - | *3A assemply of XylE, J23101 promoter and pSB1C3 vector. | + | * 3A assemply of XylE, J23101 promoter and pSB1C3 vector. |
| - | *Transformation of XL-Blue competent E.coli with the above construct. | + | * Transformation of XL-Blue competent E.coli with the above construct. |
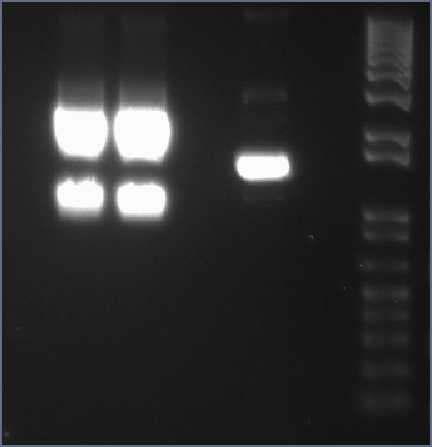
[[Image:IC_XylE-J23101-pSB1C3_Clone_Gel2.JPG|thumb|center|400px|gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation]] | [[Image:IC_XylE-J23101-pSB1C3_Clone_Gel2.JPG|thumb|center|400px|gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation]] | ||
gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation | gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation | ||
| - | *I followed Chris's Bacillus transformation protocol to transform Bacillus with constitutive GFP and RFP DNA as well as a control without DNA | + | * I followed Chris's Bacillus transformation protocol to transform Bacillus with constitutive GFP and RFP DNA as well as a control without DNA |
| - | + | '''Thursday, 19-Aug-2010''' | |
| - | *We run the XylEGFP1 PCR reaction to construct the His-GFP-Flag and Linker-XylE-Spe construct. | + | * We run the XylEGFP1 PCR reaction to construct the His-GFP-Flag and Linker-XylE-Spe construct. |
| - | + | '''Friday, 20-Aug-2010''' | |
The J23101 gene in a biobrick vector containg RFP gene | The J23101 gene in a biobrick vector containg RFP gene | ||
| - | * | + | * We run a gel on XylEGFP1 PCR reaction. Results: GFP was extended successfully, XylE extension FAILED (too much non-specific annealing) |
| - | * | + | * Catechol assay on 2hrs bench ligation of promoter,xylE and vector failed. |
| - | * | + | * Two replica plates of overnight ligated J23101, XylE and pSB1C3 transformed E.coli (one for catechol assay) |
| - | * | + | * The His-GFP-Flag DNA was gel purified |
| - | * | + | * Transformation of XL-1Blue cells with J23101 in J62001 vector from the registry. One more attemp to construct a successful promoter-xylE ligation, since we believe that the strand annealed promoter was of bad quality |
|} | |} | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Week 8 |
|- | |- | ||
| | | | ||
| + | <html> | ||
| + | <table width="850px" border="0"> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFFF66"> | ||
| + | <b>Week 8</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Monday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Tuesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Wednesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Thursday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Friday</b> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Morning</b> | ||
| + | </td> | ||
| - | = | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> |
| + | <ul> | ||
| + | <li>PCR extention of His-GFP-flag round 2</li> | ||
| + | </ul> | ||
| + | </td> | ||
| - | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | |
| + | <ul> | ||
| + | <li>midi-prep registry obtained J23101</li> | ||
| + | <li>gel purification of XbaI-His-GFP-flag-TEVs</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>gel analysis of overnight ligation and gel separation</li> | ||
| + | <li>gel analysis of linker-XylE-SpeI PCR reaction </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>make replica plate and catechol assay plate </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>catechol assay</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:50px;text-align:top;"><b>Lunch Break</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Gel purification of linker-XylE-Spe PCR construct</li> | ||
| + | <li>Gel analysis of His-GFP-flag round 2 PCR rection and then gel separation of DNA </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>restriction digestion, gel analysis and gel purification of registry obtained J23101 </li> | ||
| + | <li>PCR extension of linker-XylE-SpeI</li> | ||
| + | <li>ligation of pSB1C3, J23101 and XylE (overnight)</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>gel purification of overnight ligation</li> | ||
| + | <li>transformation of E.coli with overnight ligation product and selection by plating</li> | ||
| + | <li>gel purification of TEVs-linker-XylE-SpeI construct </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>PCR construction of fusion protein</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Gel analysis and gel purification of fusion Xyl E protein</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| - | + | '''Monday, 23-Aug''' | |
| - | * | + | * Set up overnight cultures for midi-prep |
| - | * | + | * Gel separation of linker-XylE-Spe DNA <font color = red>'''FAILED'''</font> |
| - | *PCR reaction for extension of His-GFP-Flag | + | * PCR reaction for extension of His-GFP-Flag |
| - | *Catechol assay on E.coli transformed with overnight ligated J23101, XylE and pSB1C3. <font color = red>'''FAILED'''</font> | + | * Catechol assay on E.coli transformed with overnight ligated J23101, XylE and pSB1C3. <font color = red>'''FAILED'''</font> |
| - | + | '''Tuesday, 24th-Aug''' | |
| - | * | + | * Midi-prepped registry obtained J23101. A yield of 130ng/ul of promoter was obtained. The promoter is in a biobrick vector called J62001. The promoter is upstream of RFP gene. |
*the vector carrying the promoter was digested with SpeI and PstI, while XylE gene was digested with XbaI and PstI. | *the vector carrying the promoter was digested with SpeI and PstI, while XylE gene was digested with XbaI and PstI. | ||
| - | * | + | * The promoter and the XylE gene were gel purified. |
| - | * | + | * A reaction between the promoter(still on vector) and XylE was set on for overnight ligation |
| - | *PCR purification of GFP2 -> GFP construct ready for full fusion protein. | + | * PCR purification of GFP2 -> GFP construct ready for full fusion protein. |
| - | *Gel purification of XylE lost DNA along the way. Thus PCR XylE1 with gradient for temperature scanning (taq): PCR round 1 included 62C and only rev primer to create pool of successful extensions with with rev primer (60-62). PCR round 2 with Fwd primer and temperature scale (68-68, 72-74). | + | * Gel purification of XylE lost DNA along the way. Thus PCR XylE1 with gradient for temperature scanning (taq): PCR round 1 included 62C and only rev primer to create pool of successful extensions with with rev primer (60-62). PCR round 2 with Fwd primer and temperature scale (68-68, 72-74). |
| - | + | '''Wednesday, 25th-Aug''' | |
Performed gel analysis on the purified XylE and J23101 to obtain ratios for ligation. First gel was scrapped as it produced appauling(explanation for Nick:really bad) results, 2nd gel run was successful. | Performed gel analysis on the purified XylE and J23101 to obtain ratios for ligation. First gel was scrapped as it produced appauling(explanation for Nick:really bad) results, 2nd gel run was successful. | ||
| - | *Performed a ligation reaction between the vector containing J23101, and XylE(one on bench and one overnight one). | + | * Performed a ligation reaction between the vector containing J23101, and XylE(one on bench and one overnight one). |
| - | *Transformation of the new plasmid into competent E.coli. Successfully transformed colonies can be selected for by loss of RFP expression. | + | * Transformation of the new plasmid into competent E.coli. Successfully transformed colonies can be selected for by loss of RFP expression. |
* XylE-1 PCR with temperature cascade. Gel analysis and purification. | * XylE-1 PCR with temperature cascade. Gel analysis and purification. | ||
| - | + | '''Thursday, 26th-Aug''' | |
| - | * | + | * White colonies from the promoter-XylE transformed E.coli were picked and transferred to new amp plates. One is the replica plate and the other is the catechol assay plate. |
* XylE-1, two rounds of PCR/purification were run to obtain a sufficiently clear band. An additional PCR run for XylE-1 was discarded afterwards. | * XylE-1, two rounds of PCR/purification were run to obtain a sufficiently clear band. An additional PCR run for XylE-1 was discarded afterwards. | ||
| - | + | '''Friday, 27th-Aug''' | |
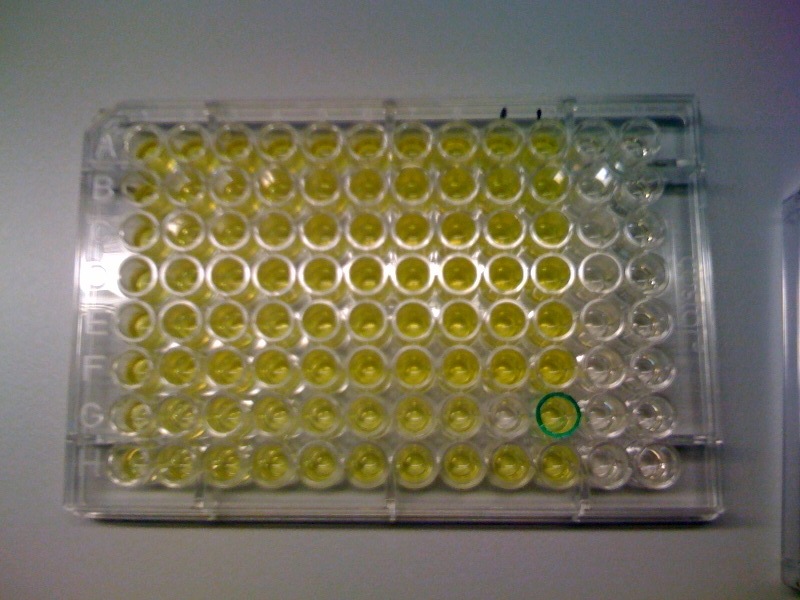
| - | *Catechol assay performed on promoter-XylE transformed E.coli. <font color = green>'''SUCCESSFULL'''</font> | + | * Catechol assay performed on promoter-XylE transformed E.coli. <font color = green>'''SUCCESSFULL'''</font> |
[[Image:IC_Catechol_Assay_before.jpg|thumb|left|Plate before adding catechol assay]] [[Image:IC_Catechol_assay_after_(27-8).jpg|thumb|center|After addition of catechol colonies turn yellow-orange in seconds!!]] | [[Image:IC_Catechol_Assay_before.jpg|thumb|left|Plate before adding catechol assay]] [[Image:IC_Catechol_assay_after_(27-8).jpg|thumb|center|After addition of catechol colonies turn yellow-orange in seconds!!]] | ||
* XylE-2 PCR and gel-purification cycles (2x) to obtain clear band. XylE-2 is now ready for assembly of the GFP-XylE fusion protein. | * XylE-2 PCR and gel-purification cycles (2x) to obtain clear band. XylE-2 is now ready for assembly of the GFP-XylE fusion protein. | ||
| - | + | '''Saturday, 28th-Aug''' | |
* Preparing the annealing step between the GFP-2 and XylE-2 constructs, we discovered sequence dissimilarities in the TEV-cleavable regions which we planned to use for the annealing step. Nevertheless a PCR was run with appropriate conditions (allowing for a minimal amount of unspecific annealing). | * Preparing the annealing step between the GFP-2 and XylE-2 constructs, we discovered sequence dissimilarities in the TEV-cleavable regions which we planned to use for the annealing step. Nevertheless a PCR was run with appropriate conditions (allowing for a minimal amount of unspecific annealing). | ||
| - | + | '''Sunday, 29th-Aug''' | |
* Gel analysis of the attempted annealing reaction of GFP-2 XylE-2 showed unsufficiently clear bands for gel-purification. A new reaction is being prepared: 10 rounds of annealing PCR, followed by addition of primers (5' primer for GFP-2 and 3' primer for XylE-2) in order to introduce an amplification step in the reaction. --- Following Kirstins advice, we are discarding this reaction and wait for the arrival of new primers for XylE-2 (5' + TEV) and GFP-2 (3' +TEV). | * Gel analysis of the attempted annealing reaction of GFP-2 XylE-2 showed unsufficiently clear bands for gel-purification. A new reaction is being prepared: 10 rounds of annealing PCR, followed by addition of primers (5' primer for GFP-2 and 3' primer for XylE-2) in order to introduce an amplification step in the reaction. --- Following Kirstins advice, we are discarding this reaction and wait for the arrival of new primers for XylE-2 (5' + TEV) and GFP-2 (3' +TEV). | ||
| Line 292: | Line 495: | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Week 9 |
|- | |- | ||
| | | | ||
| + | <html> | ||
| + | <table width="850px" border="0"> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFFF66"> | ||
| + | <b>Week 9</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Monday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Tuesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Wednesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Thursday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Friday</b> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Morning</b> | ||
| + | </td> | ||
| - | = | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> |
| + | <ul> | ||
| + | <li>mini-prep of promoter-XylE transformed E.col</li> | ||
| + | <li>design of reverse primer for GFP to add the corrected TEV sequence to the construct.</li> | ||
| + | </ul> | ||
| + | </td> | ||
| - | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | |
| + | <ul> | ||
| + | <li>midi-prep of promoter-XylE transformed E.coli</li> | ||
| + | <li>restriction digestion of pSB1C3+terminator vector DNA</li> | ||
| + | <li>restriction digestion of promoter-XylE DNA </li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Gel purification kit of cut promoter-XylE DNA</li> | ||
| + | <li>Preparation of M9 and LB medium for assay </li> | ||
| + | <li>Gel analysis of of insert and vector DNA</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>find optimun wavelength for catechol assays</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>find optimun cell density and catechol concentration for assay</li> | ||
| + | <li>Reverse primer for GFP with modified TEV arrived - primer dilution</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:50px;text-align:top;"><b>Lunch Break</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>gel analysis of mini-prepped promoter-XylE</li> | ||
| + | <li>cross-check for primer design (GFP-TEV-2) and ordering</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>PCR purification of cut pSB1C3+terminator vector DNA </li> | ||
| + | <li>Gel analysis of cut promoter-XylE DNA</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>dephosphorylation of the vector+terminator</li> | ||
| + | <li>ligation reaction between promoter-XylE insert and pSB1C3+terminator vector</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>GFP 2 PCR with new reverse primer (adopted TEV sequence)</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| - | + | '''Monday, 30th-Aug''' | |
* Mini prep of 4 x J23101-XylE taken from 4 different colonies (8 14 24 27). | * Mini prep of 4 x J23101-XylE taken from 4 different colonies (8 14 24 27). | ||
* The gel analysis showed successful vector uptake. | * The gel analysis showed successful vector uptake. | ||
| Line 307: | Line 614: | ||
* A new primer was designed in order to add a corrected TEV-protease-cleavable sequence to the His-GFP-Flag construct. This was controlled and ordered. | * A new primer was designed in order to add a corrected TEV-protease-cleavable sequence to the His-GFP-Flag construct. This was controlled and ordered. | ||
| - | + | '''Tuesday, 31st-Aug''' | |
* Midi prep of colony 24 for XylE-J23101 the final concentration was ~100ng/ul which wasnt so great but Chris says the protocol produces very poor yields. | * Midi prep of colony 24 for XylE-J23101 the final concentration was ~100ng/ul which wasnt so great but Chris says the protocol produces very poor yields. | ||
* We are performing the next building step of our vector. PSB1C3 containing terminator B0014 was cut with EcoRI and XbaI. The insert was cut with EcoRI and SpeI and both were incubated for 1.5hrs. Wolf is now running a gel to purify out the insert via gel purification and perform a PCR purification on the vector. | * We are performing the next building step of our vector. PSB1C3 containing terminator B0014 was cut with EcoRI and XbaI. The insert was cut with EcoRI and SpeI and both were incubated for 1.5hrs. Wolf is now running a gel to purify out the insert via gel purification and perform a PCR purification on the vector. | ||
| Line 313: | Line 620: | ||
*We shall see purification expert Kieran tomorrow and talk through the process. Chris will also talk us through our characterization of XylE experiments- we will use the robot after it's been programmed but until then we can use the plate-reader. | *We shall see purification expert Kieran tomorrow and talk through the process. Chris will also talk us through our characterization of XylE experiments- we will use the robot after it's been programmed but until then we can use the plate-reader. | ||
| - | + | '''Thursday, 2nd-Sept''' | |
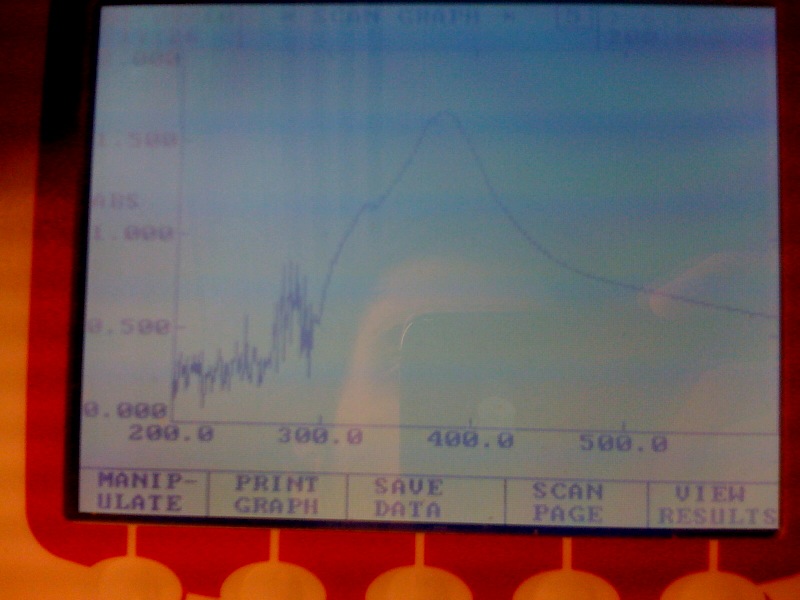
[[Image:IC_200-600nm_spectr.jpg|thumb||center|300px|Spectra of XylE transformed E.coli after addition of catechol assay. The '''broad peak around 380nm wavelength''' arises is due to the presence of the product of the enzymatic reaction involving pyrocatechol and XylE enzyme. This peak if absent if a culture of XylE transformed cells are measured without the addition of catechol]] | [[Image:IC_200-600nm_spectr.jpg|thumb||center|300px|Spectra of XylE transformed E.coli after addition of catechol assay. The '''broad peak around 380nm wavelength''' arises is due to the presence of the product of the enzymatic reaction involving pyrocatechol and XylE enzyme. This peak if absent if a culture of XylE transformed cells are measured without the addition of catechol]] | ||
*Spectrophotometry experiments with XylE transformed E.coli in LB medium (M9 culture was contaminated) reveiled the followings: On catechol assay of the trasformed cells, the '''positive yellow output can be quantitively measured by a broad peak at 380nm.''' | *Spectrophotometry experiments with XylE transformed E.coli in LB medium (M9 culture was contaminated) reveiled the followings: On catechol assay of the trasformed cells, the '''positive yellow output can be quantitively measured by a broad peak at 380nm.''' | ||
*Trasformation of competent E.coli cells with promoter-XylE-terminator pSB1C3vector. | *Trasformation of competent E.coli cells with promoter-XylE-terminator pSB1C3vector. | ||
| - | + | '''Friday, 3rd-Sept''' | |
*experiment to determine concentrations of catechol and cell density for assays | *experiment to determine concentrations of catechol and cell density for assays | ||
| Line 328: | Line 635: | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Week 10 |
|- | |- | ||
| | | | ||
| - | == | + | <html> |
| + | <table width="850px" border="0"> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFFF66"> | ||
| + | <b>Week 10</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Monday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Tuesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Wednesday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Thursday</b> | ||
| + | </td> | ||
| + | <td style="background-color:#FFFF99;"><b>Friday</b> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Morning</b> | ||
| + | </td> | ||
| - | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | |
| + | <ul> | ||
| + | <li>PCR extension of Pveg promoter </li> | ||
| + | </ul> | ||
| + | </td> | ||
| - | = | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> |
| - | + | <ul> | |
| + | <li>Gel purification of extended Pveg promoter</li> | ||
| + | <li>Annealing PCR for GFP-XylE fusion protein</li> | ||
| + | </ul> | ||
| + | </td> | ||
| - | *I performed a catechol assay on the picked transformed colonies to deduce which ones were successfully transformed with the insert plus vector. 1-5 7 and 10 failed to turn yellow (1-5 were background controls)leaving 8 yellow colonies. | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> |
| - | *I had to perform a colony PCR on two selected colonies 8 and 14 to check the correct insert+terminator was present. | + | <ul> |
| - | *Colony 8 when run on a gel analysis showed the correct size 924 XylE + 97 J23101 + 35 B0014. | + | <li>vector with insert tranformed into E.coli and plate to select for successful transformation</li> |
| - | *This was set up as an overnight culture. | + | </ul> |
| - | *Gel analysis of the GFP-TEV construct showed satisfactory bands. | + | </td> |
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>prepare overnight cultures of pVeg-RBS transformed E.coli </li> | ||
| + | <li>Gel-analysis of amplification-PCR for XylE-2</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>midi-prep of Pveg-RBS vector</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:50px;text-align:top;"><b>Lunch Break</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:50px;width:200px;text-align:top;"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Gel analysis and purification of the GFP-TEV construct</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>restriction digestion and gel purification of thr extended Pveg promoter</li> | ||
| + | <li>overnight ligation of the cut Pveg promoter in a vector </li> | ||
| + | <li>Gel analysis of GFP-XylE annealing PCR - unsuccessful reaction</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Amplification PCR for XylE-2</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | <li>Gel-purification of amplification-PCR for XylE-2</li> | ||
| + | </ul> | ||
| + | </td> | ||
| + | |||
| + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
| + | <ul> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | '''Monday, 6th-Sept''' | ||
| + | * PCR extension of Pveg promoter: EcoRI---Pveg---RBS-SpeI | ||
| + | |||
| + | * I performed a catechol assay on the picked transformed colonies to deduce which ones were successfully transformed with the insert plus vector. 1-5 7 and 10 failed to turn yellow (1-5 were background controls)leaving 8 yellow colonies. | ||
| + | * I had to perform a colony PCR on two selected colonies 8 and 14 to check the correct insert+terminator was present. | ||
| + | * Colony 8 when run on a gel analysis showed the correct size 924 XylE + 97 J23101 + 35 B0014. | ||
| + | * This was set up as an overnight culture. | ||
| + | * Gel analysis of the GFP-TEV construct showed satisfactory bands. | ||
| - | + | '''Tuesday 7th Sept''' | |
| - | *Mini prep of the overnight Colony 8 culture (by Wolf) | + | * Mini prep of the overnight Colony 8 culture (by Wolf) |
| - | *Midi prep of overnight culture | + | * Midi prep of overnight culture |
| - | * | + | * Gel purification of PCR product EcoRI--Pveg--RBS--SpeI |
| - | * | + | * Overnight restriction digestion of EcoRI--Pveg--RBS--SpeI with SpeI |
| - | * | + | * Started data analysis of plate assay |
| - | *Annealing PCR reaction included 2 samples without and one with additional primers (XbaI-His-GFP Fwd, SpeI-XylE Rev). While the primer-including reaction did not show any clearly identifiable bands, the others showed clear, if very weak bands at 800 and 1000bp, which represent the GFP and XylE constructs respectively. No band was identifiable at the 1.7 kb range, which would have indicated a successful annealing reaction. However, problems with the lense of the gel-analyser were only discovered later to have severely reduced the band brightness. Potentially a PCR-reaction with appropriate primers to amplify an annealed product(XbaI-His-GFP Fwd, SpeI-XylE Rev), could have been successful. However the reaction mixture was disposed of before this became clear. | + | * Annealing PCR reaction included 2 samples without and one with additional primers (XbaI-His-GFP Fwd, SpeI-XylE Rev). While the primer-including reaction did not show any clearly identifiable bands, the others showed clear, if very weak bands at 800 and 1000bp, which represent the GFP and XylE constructs respectively. No band was identifiable at the 1.7 kb range, which would have indicated a successful annealing reaction. However, problems with the lense of the gel-analyser were only discovered later to have severely reduced the band brightness. Potentially a PCR-reaction with appropriate primers to amplify an annealed product(XbaI-His-GFP Fwd, SpeI-XylE Rev), could have been successful. However the reaction mixture was disposed of before this became clear. |
| - | + | '''Wednesday, 8th Sep''' | |
| - | *A second PCR extension of Pveg promoter to introduce the RBS and cut sites. It was also gel purified and stored in gel lumbs in the freezer. (maybe needed later so keep in mind). | + | * A second PCR extension of Pveg promoter to introduce the RBS and cut sites. It was also gel purified and stored in gel lumbs in the freezer. (maybe needed later so keep in mind). |
| - | *Overnight restriction digestion completed. Then, it was run on the gel to check if it worked and then gel purified again. | + | * Overnight restriction digestion completed. Then, it was run on the gel to check if it worked and then gel purified again. |
| - | *Vector PSBI-C3 was digested to remove the terminator and make it sticky for the insert (Pveg-RBS). Then it was run on the gel to check if restriction has worked, but the gel didn't run far enough to determine easily between undigested and digested vector. | + | * Vector PSBI-C3 was digested to remove the terminator and make it sticky for the insert (Pveg-RBS). Then it was run on the gel to check if restriction has worked, but the gel didn't run far enough to determine easily between undigested and digested vector. |
* For further annealing reactions for GFP-XylE constructs additional XylE(2) template was required -> amplification PCR for XylE(2). | * For further annealing reactions for GFP-XylE constructs additional XylE(2) template was required -> amplification PCR for XylE(2). | ||
| - | + | '''Thursday, 9th Sep''' | |
| - | *Gel with digested and undigested vector PSBI-C3 was run and then the digested vector was purified. | + | * Gel with digested and undigested vector PSBI-C3 was run and then the digested vector was purified. |
| - | *Gel was run to determine the DNA concentration ratio for the ligation of PSBI-C3 and Pveg-RBS. | + | * Gel was run to determine the DNA concentration ratio for the ligation of PSBI-C3 and Pveg-RBS. |
| - | *Vector PSBI-C3 was dephosphorylated. | + | * Vector PSBI-C3 was dephosphorylated. |
| - | *Ligation of PSBI-C3 and Pveg-RBS has been set up overnight. | + | * Ligation of PSBI-C3 and Pveg-RBS has been set up overnight. |
| - | *Gel-analysis and gel-purification of the XylE(2) amplification PCR product. | + | * Gel-analysis and gel-purification of the XylE(2) amplification PCR product. |
| - | *The transformation of E.Coli with Pveg-RBS in PSBI-C3 and PSBI-C3 by itself (to check see how successful dephosphoryaltion of PSBI-C3 was and estimate the percentage of bacteria that contain the insert) was completed | + | * The transformation of E.Coli with Pveg-RBS in PSBI-C3 and PSBI-C3 by itself (to check see how successful dephosphoryaltion of PSBI-C3 was and estimate the percentage of bacteria that contain the insert) was completed |
| - | * | + | * Concentration of the midi prep of J23101-XylE-B0014 was determined to be ~600ng/ul (using new protocol) |
| - | *Kyasha kindly digested my midi with EcoRI and SpeI and performed a gel analysis. The results show a potential additional plasmid contaminating my midi however the concentration of DNA was extremely high. NB Chris said that it could be sheared DNA from a midi prep step. | + | * Kyasha kindly digested my midi with EcoRI and SpeI and performed a gel analysis. The results show a potential additional plasmid contaminating my midi however the concentration of DNA was extremely high. NB Chris said that it could be sheared DNA from a midi prep step. |
| - | * | + | * The midi prepped plasmid was transformed into testing E.coli strain TOP10. |
| - | + | '''Friday, 10 sep''' | |
The transformation was a SUCCESS. 2x replica plates were made plate 1# 1-6 plate #2 6-11; colony 6 and colony 9 of plates 1 and 2 respectively were transfered into a 5ml liquid culture + 5ul CmR. These will later be turned into glycerol stocks. After the replica plates have grown up mini preps on a number of colonies shall be performed - this hopefully will eliminate the contaminating plasmid DNA. This will be followed by a midi prep. | The transformation was a SUCCESS. 2x replica plates were made plate 1# 1-6 plate #2 6-11; colony 6 and colony 9 of plates 1 and 2 respectively were transfered into a 5ml liquid culture + 5ul CmR. These will later be turned into glycerol stocks. After the replica plates have grown up mini preps on a number of colonies shall be performed - this hopefully will eliminate the contaminating plasmid DNA. This will be followed by a midi prep. | ||
| - | + | '''Saturday, 11 Sep''' | |
The transformation of E.Coli with PSB1-C3 with insert did not work :( | The transformation of E.Coli with PSB1-C3 with insert did not work :( | ||
| Line 382: | Line 791: | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| - | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"| | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Week 11 |
|- | |- | ||
| | | | ||
| - | + | '''Monday 13th september''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Overnights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. | Overnights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. | ||
Revision as of 22:34, 24 October 2010
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
| Team XylE |
Follow our progress: Click me!
| XylE team Lab Objectives |
|
| Week 6 | ||||||||||||||||||||||||
|
we constructed the standard E.coli promoter J23101 with sticky ends. These ends are complementary to restriction sites made by EcoRI and SpeI enzyme. This promoter will be later used in 3A assemply to construct a promoter-RBS-XylE design in a psB1C3 vector. E.coli will be transformed with this final construct plasmid to assess XylE activity and characterization. It will also be one of the submitted biobricks.
these cultures are going to be used tomorrow for mini-prepping. Miniprep will allow us to isolate E.coli's plasmid DNA(which contains the XylE gene). Friday, 13-Aug-2010
Mini-prep is usually used to confirm that our gene of interest has not been changed in any way, as the isolated plasnid id sent for sequencing. However, since XylE was taken from the registry, we assume that it is fine and no sequencing is required. The mini-prep will later be used for the midi-prep (that gives out higher yeilds of DNA needed for cloning).
|
| Week 7 | ||||||||||||||||||||||||
|
Tuesday, 17-Aug-2010
gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation
Thursday, 19-Aug-2010
Friday, 20-Aug-2010 The J23101 gene in a biobrick vector containg RFP gene
|
| Week 8 | ||||||||||||||||||||||||
|
Monday, 23-Aug
Tuesday, 24th-Aug
Wednesday, 25th-Aug Performed gel analysis on the purified XylE and J23101 to obtain ratios for ligation. First gel was scrapped as it produced appauling(explanation for Nick:really bad) results, 2nd gel run was successful.
Thursday, 26th-Aug
Friday, 27th-Aug
Saturday, 28th-Aug
Sunday, 29th-Aug
|
| Week 10 | ||||||||||||||||||||||||
|
Tuesday 7th Sept
Wednesday, 8th Sep
Thursday, 9th Sep
Friday, 10 sep The transformation was a SUCCESS. 2x replica plates were made plate 1# 1-6 plate #2 6-11; colony 6 and colony 9 of plates 1 and 2 respectively were transfered into a 5ml liquid culture + 5ul CmR. These will later be turned into glycerol stocks. After the replica plates have grown up mini preps on a number of colonies shall be performed - this hopefully will eliminate the contaminating plasmid DNA. This will be followed by a midi prep. Saturday, 11 Sep The transformation of E.Coli with PSB1-C3 with insert did not work :( |
| Week 11 |
|
Monday 13th september Overnights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. Tues 14th September
The assay was carried out with E.coli, top ten spcies, transformed with J23101-XylE-B0014 in pSB1C3 vector.The overnight culture wastransfered in new medium this morning for 4 hrs before assaying. LB medium was used for dilutions and blank. Catechol was diluted in ddH2O. Data analysis of the assay
Piotr:
Weds 15th September
Thurs 16th September
- digest with XbaI and SpeI - PCR reactions with the following primers added: HIS-GFP, XylE-Xba and the other sample with HIS-GFP and GFP-flag The PCR reactions were not very conclusive on the gels but digests allowed to determine that colonies 1->5 seem to have the right sizes of DNA in them and on Friday they will be prepared to be sent off for sequencing
Friday 17th September
The gel purifications of 3k3 vector and XylE were used for ligation. 3K3 was dephosphorylated and then ligated in a ratio 5:1 with the insert. This was left overnight and Chris tried to transform with it on sunday.. ligation and transformation failed :-( |
| Week 12 |
Monday 20th SeptPerformed ligation again. Errors in the sequencing we received back regarding J23101-XylE-B0014. One was a log error (in XylE) phew. The other is in PSB1C3 out of the scar site and should be OK. Chris is currently assembling the pVEG promoter+ RBS in a vector. Once he has finished this, I will combine XylE-GFP and XylE with this promoter for comparison characterization with J23101 (in E.coli and Bacillus) Tues 21st Sept
Wed 22nd Sept
Thurs 23rd Sept First day of AUTUMN!!
Fri 24th Sept
|
| Week 13 |
Monday 27th Sept
Tues 28th Sept
Weds 29th Sept
Thurs 30th Sept
Friday 1st Oct
ahahahaha awesome. hey you :)
|
have a look :)it's from John Hoppkins wiki 2008. There are one or two good ones! Love: Before I heard the doctors tell The dangers of a kiss; I had considered kissing you. The nearest thing to bliss. But now I know biology and sit and sigh and moan; six million mad bacteria and I thought we were alone!
|
| Week 14 |
Monday 4th OctDid a replica plate and catechol assay plate/ minis 2 6 7 9 12 16. (Successful Pveg-XylE transformation) Tues 5th Oct4 16 colonies were background the rest turned yellow. Mini prepped 2 6 7 9 12 16. Made overnight assay cultures of 3k3-XylE and C3-xylE Weds 6th Oct
Thursday 7th Oct
Friday 8th Oct
Sunday 10th of the 10th of the 2010!!!!! and im in LAB
|
| Week 15 |
Monday 11th Oct
Tues 12th Oct
Weds 13th Octmini prepped 3k3-Pveg-XylE-B0014 and went to the school workshop! set off overnight midi of culture 9 Thurs 14th OctMidi prepped 3k3-Pveg Fri 15thTransformed all my constructs into T0P10 3k3-J23101/pveg and PSB1C3-J23101/PVEG |
 "
"