Team:SDU-Denmark/project-r
From 2010.igem.org
(→Biobrick assembly and sequence) |
(→Biobrick assembly and sequence) |
||
| Line 442: | Line 442: | ||
== Retinal == | == Retinal == | ||
=== Biobrick assembly and sequence === | === Biobrick assembly and sequence === | ||
| - | We succesfully assembled the biobrick K343006, which consists of the TetR repressible promoter R0040 followed by the ribosome binding site B0034, the coding sequence K343002 and the terminators B0010 and B0012.<br> | + | We succesfully assembled the biobrick [http://partsregistry.org/Part:BBa_K343006 K343006], which consists of the TetR repressible promoter R0040 followed by the ribosome binding site B0034, the coding sequence K343002 and the terminators B0010 and B0012.<br> |
This was achieved through "standard" silver assembly and confirmed via sequencing with biobrick verification forward 2 (VF2) and verification reverse (VR) primers.<br> | This was achieved through "standard" silver assembly and confirmed via sequencing with biobrick verification forward 2 (VF2) and verification reverse (VR) primers.<br> | ||
Sequencing results (as .ab1 files):<br> | Sequencing results (as .ab1 files):<br> | ||
Revision as of 19:30, 24 October 2010
Results
Photosensor
Biobrick assembly and sequence
We succesfully assembled the biobrick [http://partsregistry.org/Part:BBa_K343007 K343007], which consists of the TetR repressible promoter R0040 followed by the ribosome binding site B0034, the coding sequence K343003 and the terminators B0010 and B0012.
This was achieved through "standard" silver assembly and confirmed via sequencing with biobrick verification forward 2 (VF2) and verification reverse (VR) primers.
Sequencing results (as .ab1 files):
Sequencing results K343007
When comparing the sequence that came back from sequencing with the theoretical sequence the part should have it is identical, except for a silent mutation in the coding sequence, which does not change the amino acid sequence.
Effect
Our results from the experiments with semi-solid agar confirm that the biobrick does indeed couple itself to the bacterial chemotaxis pathway and modify the bacterial motility pattern by modifying the tumbling frequency. For further information on this conclusion see Characterization - K343007 - 1. Growth of bacterial culture on semi-solid agar plates (Experiment 1 and 2)
The videomicroscopy indicates that blue light with a wavelength around 500nm leads to CheA's autophosphorylation being downregulated. This means that the bacterial tumbling frequency gets reduced and the bacteria will spend an increased time in the "run" mode of propulsion. This means that the bacteria will travel further in a certain amount of time, compared to wildtype bacteria. More details can be found here Characterization - K343007 - 2. Videomicroscopy and computer analysis of bacterial motility
Stability assay
| Stability assay. | ||||
| 0 generations | Dilution number | Colonies counted on plate | Cfu | PS-chlor/PS+chlor |
| PS1 + chlor | 6 | 168 | 8.4E+09 | 100 |
| PS1 - chlor | 6 | 157 | 7.85E+09 | |
| PS2 + chlor | 6 | 192 | 9.6E+09 | 100 |
| PS2 - chlor | 6 | 216 | 1.08E+10 | |
| 20 generations | ||||
| PS1 + chlor | 6 | 157 | 7.85E+09 | 59.24528302 |
| PS1 - chlor | 6 | 265 | 1.325E+10 | |
| PS2 + chlor | 6 | 130 | 6.5E+09 | 63.41463415 |
| PS2 - chlor | 6 | 205 | 1.025E+10 | |
| 40 generations | ||||
| PS1 + chlor | 6 | 143 | 7.15E+09 | 95.97315436 |
| PS1 - chlor | 6 | 149 | 7.45E+09 | |
| PS2 + chlor | 6 | 168 | 8.4E+09 | 92.81767956 |
| PS2 - chlor | 6 | 181 | 9.05E+09 | |
| 60 generations | ||||
| PS1 + chlor | 6 | 147 | 7.35E+09 | 89.63414634 |
| PS1 - chlor | 6 | 164 | 8.2E+09 | |
| PS2 + chlor | 6 | 134 | 6.7E+09 | 104.6875 |
| PS2 - chlor | 6 | 128 | 6.4E+09 | |
| 80 generations | ||||
| PS1 + chlor | 6 | 127 | 6.35E+09 | 136.5591398 |
| PS1 - chlor | 6 | 93 | 4.65E+09 | |
| PS2 + chlor | 6 | 99 | 4.95E+09 | 104.2105263 |
| PS2 - chlor | 6 | 95 | 4.75E+09 | |
| 100 generations | ||||
| PS1 + chlor | 5 | 130 | 650000000 | 125 |
| PS1 - chlor | 5 | 104 | 520000000 | |
| PS2 + chlor | 5 | 124 | 620000000 | 118.0952381 |
| PS2 - chlor | 5 | 105 | 525000000 | |
| Number of generations | PS1 | PS2 | PS (average) | Standard deviations |
| 0 | 100 | 100 | 100 | 0 |
| 20 | 59.24528 | 63.41463 | 61.32995858 | 2.948176455 |
| 40 | 95.97315 | 92.81768 | 94.39541696 | 2.231257632 |
| 60 | 89.63415 | 104.6875 | 97.16082317 | 10.64432845 |
| 80 | 136.5591 | 104.2105 | 120.3848331 | 22.87392395 |
| 100 | 125 | 118.0952 | 121.547619 | 4.882403965 |
Retinal
Biobrick assembly and sequence
We succesfully assembled the biobrick [http://partsregistry.org/Part:BBa_K343006 K343006], which consists of the TetR repressible promoter R0040 followed by the ribosome binding site B0034, the coding sequence K343002 and the terminators B0010 and B0012.
This was achieved through "standard" silver assembly and confirmed via sequencing with biobrick verification forward 2 (VF2) and verification reverse (VR) primers.
Sequencing results (as .ab1 files):
Sequencing results for K343006
When comparing the theoretical sequence with the sequence done on the part K343006 the two sequences was identical.
Effect
Stability assay
Flagella
Motility assay
Exp. 1
The purpose of this experiment is to see if the cells containing pSB1C3-K343004 move farther than the wild type (MG1655) and the negative control (DH5alpha). This would be an indication of hyperflagellation. We also want to test if it makes a difference in the motility wether the bacteria contain low-medium- or high-copy plasmids.
For the motility experiments we added 5ul of an ON culture to petridishes containing motility agar (LB media with 0.3% agar) instead of regular LA (Luria agar). This semi-solid media lets the bacteria swim more easily.
The plates were incubated at 37 degrees celcius for 24 hours.
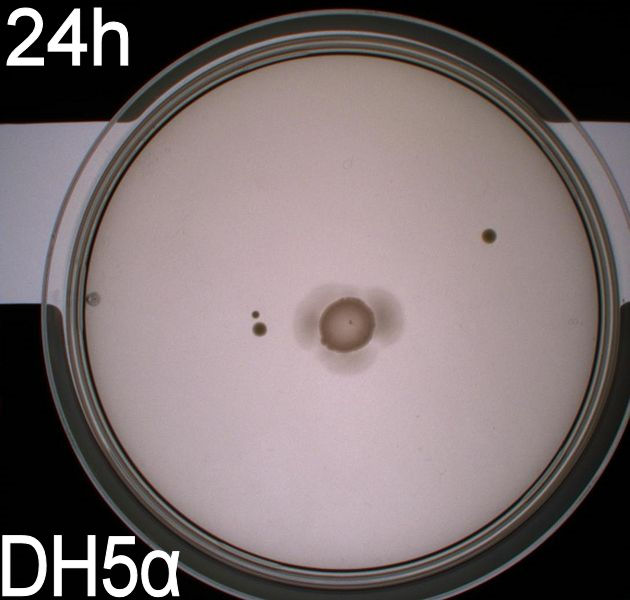


The upper two plates did not contain antibiotics, and therefore contamination colonies are seen.
The upper left picture is of E. coli strain DH5alpha that does not express flagella and therefor movement in the media should, as seen on the picture, be minimal. The upper right picture is of the wild type E. coli strain MG1655, this strain has about 8-10 flagella per cell. These cells are, as seen, expected to move farther than the DH5alpha but not as far as the transformed cells. The lower left picture is of E. Coli strain MG1655 with pSB1C3-K343004 this shows that these bacteria move farther than the wild type and the negative control. The lover right picture is of E. coli strain MG1655 with pSB3K3-K343004 these bacteria move farther than the wild type, the negative control and MG1655 with pSB3K3-K343004.
From the pictures above we can definately se that the bacteria containing our part is much more motile than the wild type. We assume this is caused by overexpression of the FlhDC master flagella operon which leads to hyperflagellation of the cells.
The two buttom pictures show that bacteria with pSB1C3-K343004 have not moved as far as the bacteria containing pSB3K3-K343004. pSB1C3 is a high copy plasmid while pSB3K3 is a low-medium copy plasmid. The promoters in K343004 is a constitutive promoter (tetR repressable promoter). Bacteria containing a high copy plasmid with a constitutive promoter are more metabolically challanged than bacteria containing a low- or medium-copy plasmid with a constitutive promoter because of the higher number of plasmids per the cell. Therefore the high copy plasmid bacteria are less motile than low- or medium-copy plasmid bacteria.
OD measurment of MG1655
| Mg1655 | PS in pSB3T5 | PS in pSB1C3 | flhDC in pSB3K3 | flhDC in pSB1C3 | NinaB in pSB1C3 | |||||||
| Hours | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 |
| 0 | 0.024 | 0.026 | 0.028 | 0.03 | 0.024 | 0.024 | 0.028 | 0.027 | 0.028 | 0.027 | 0.027 | 0.027 |
| 1 | 0.082 | 0.03 | 0.058 | 0.06 | 0.059 | 0.063 | 0.069 | 0.077 | 0.052 | 0.051 | 0.084 | 0.072 |
| 2.5 | 0.58 | 0.65 | 0.42 | 0.45 | 0.48 | 0.44 | 0.52 | 0.6 | 0.38 | 0.34 | 0.31 | 0.54 |
| 3 | 1.14 | 1.16 | 0.99 | 0.91 | 0.94 | 0.94 | 1.06 | 1.16 | 0.84 | 0.78 | 1.04 | 1.03 |
| 3.83 | 1.54 | 1.66 | 1.52 | 1.31 | 1.38 | 1.37 | 1.54 | 1.72 | 1.33 | 1.25 | 1.91 | 1.45 |
| 5 | 4.4 | 4.3 | 3.2 | 3.4 | 4.2 | 3.3 | 4.2 | 2.2 | 3.9 | 4 | 3.9 | 3.9 |
| 6 | 4.7 | 4.7 | 2.8 | 4.3 | 4.2 | 3.1 | 4.7 | 4.4 | 3.8 | 5 | 3.4 | 4.7 |
| 7 | 4 | 3.8 | 3.3 | 3.8 | 4.2 | 3.2 | 4.3 | 4.7 | 4.9 | 4.5 | 3.7 | 3.9 |
| 8 | 5.5 | 6.1 | 3.9 | 4 | 3.5 | 6.8 | 4.2 | 4.8 | 5.4 | 5.3 | 4.6 | 4.6 |
| 9 | 4.1 | 4.1 | 3.4 | 2.6 | 3.6 | 4.7 | 2.6 | 3.1 | 3.7 | 3.1 | 2.6 | 3.6 |
| 10 | 4.6 | 6.1 | 5 | 4.8 | 7.8 | 4.5 | 5 | 6 | 5.3 | 5.3 | 4.4 | 4.7 |
| 11 | 5.1 | 6.2 | 4.7 | 4 | 6.1 | 7.1 | 6.6 | 8.3 | 6.6 | 6.4 | 6.2 | 4.1 |
| 12 | 5.1 | 5 | 3 | 4.4 | 4.4 | 3.9 | 4 | 4.8 | 5 | 5.3 | 4 | 4.7 |
| 24 | 5.2 | 5.6 | 3.3 | 5.4 | 6.4 | 5.8 | 4.6 | 6.4 | 7.1 | 7.4 | 5.6 | 5.2 |
This is where it gets really interesting. Just look what we've made!
Contents |
 "
"
