Team:St Andrews/project/modelling/model 4
From 2010.igem.org
Patrickolden (Talk | contribs) |
Patrickolden (Talk | contribs) |
||
| Line 11: | Line 11: | ||
The primary development from previous models - the change of focus from one cell to the whole culture - was implemented by making many small adjustments to the previous differential equations we used. In addition to the biochemical reactions changing chemical concentrations, we also had a volumetric term describing the increase of volume (and subsequent decrease in concentration) as new cells were added to the colony. This can be understood by taking the derivative of the HSL concentration: | The primary development from previous models - the change of focus from one cell to the whole culture - was implemented by making many small adjustments to the previous differential equations we used. In addition to the biochemical reactions changing chemical concentrations, we also had a volumetric term describing the increase of volume (and subsequent decrease in concentration) as new cells were added to the colony. This can be understood by taking the derivative of the HSL concentration: | ||
| - | [[Image:4line1.gif| | + | [[Image:4line1.gif|300px]] |
<math>d[HSL]/dt = dN/dt * (1 / V) - 1 / (V ^ 2) * N </math> | <math>d[HSL]/dt = dN/dt * (1 / V) - 1 / (V ^ 2) * N </math> | ||
Revision as of 19:23, 24 October 2010


Model 4: Pseudo Multi Cell Two Dimensions
Theory & assumptions
Although our previous work presented many insights into the LuxR quorum sensing system, a new model of further complexity was deemed necessary to investigate some of the subtleties of bistable behaviour. This was accomplished by changing focus from the interactions in one cell, to the population of bacteria as a whole. In doing this, not only were we able to reproduce the results from previous models on a population-wide scale, but our results would be empirically verifiable in the lab.
The primary development from previous models - the change of focus from one cell to the whole culture - was implemented by making many small adjustments to the previous differential equations we used. In addition to the biochemical reactions changing chemical concentrations, we also had a volumetric term describing the increase of volume (and subsequent decrease in concentration) as new cells were added to the colony. This can be understood by taking the derivative of the HSL concentration:
<math>d[HSL]/dt = dN/dt * (1 / V) - 1 / (V ^ 2) * N </math>
Similar to model 3, the cell density was initially increased by a cell growth function, where the number of bacteria doubled every 20 minutes. Having ran to a realistic maximum cell density, there was a stationary phase in the cell growth to allow steady state concentrations of chemicals to be realised. The cell death function was then called, and the cell density was ran back to it's starting value.
Results
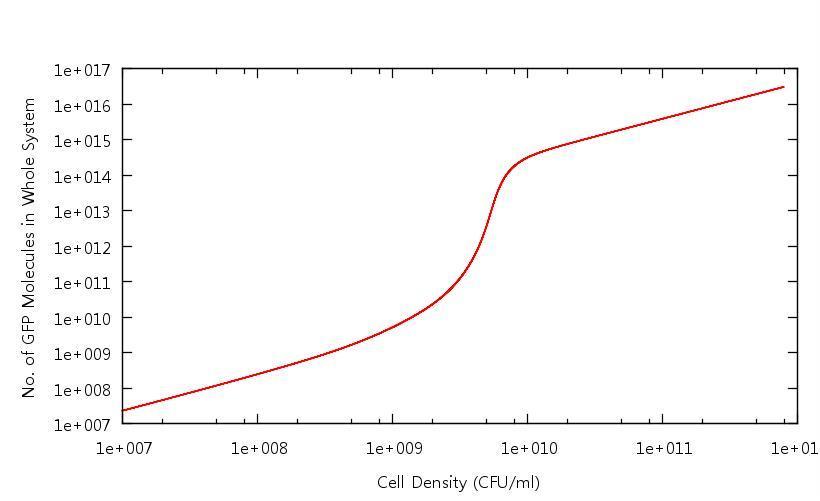
Our results show that in accordance with our single cell models, there is clear switching behaviour in the LuxR system. One point of interest is how the cell density at which the system becomes "up regulated" has changed from 10^6 CFU/ml to 10^9 CFU/ml. Tests have been carried out to confirm that this is related to the "diffusion constant" which describes the extent to which the cross-membrane concentration gradient of the signalling molecule (HSL) effects the rate of flow of HSL out of the cell. This shifted activation point can be seen below:
 "
"