Team:Imperial College London/Chassis
From 2010.igem.org
| Line 29: | Line 29: | ||
|- | |- | ||
| - | ! Required components !! Features of ''B. subtilis' | + | ! scope="col" width="225" | Required components !! scope="col" width="225" | Features of ''B. subtilis' |
|- | |- | ||
|For our '''detection module''' we need: | |For our '''detection module''' we need: | ||
| Line 54: | Line 54: | ||
'''Notes:''' | '''Notes:''' | ||
| + | |||
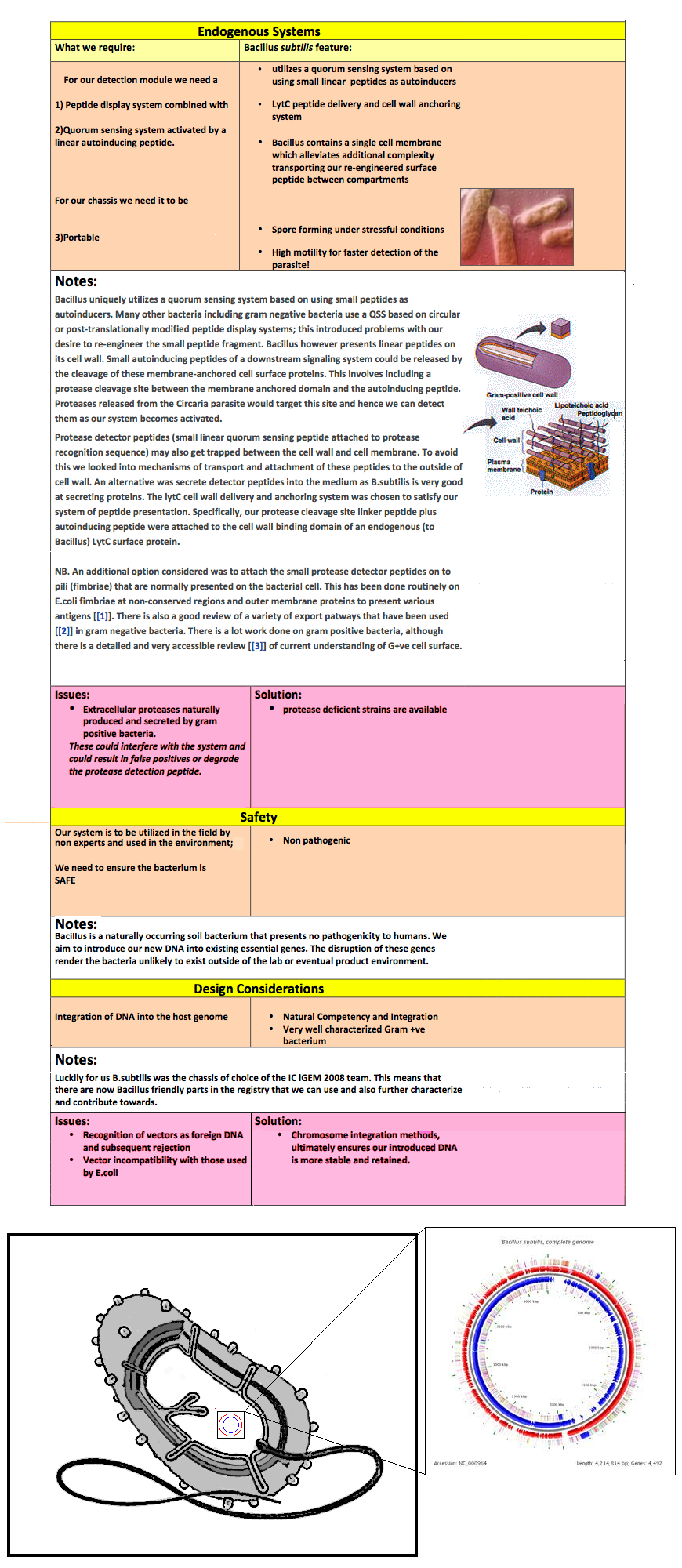
Bacillus uniquely utilizes a quorum sensing system based on using small peptides as autoinducers. Many other bacteria including gram negative bacteria use a QSS based on circular or post-translationally modified peptide display systems; this introduced problems with our desire to re-engineer the small peptide fragment. Bacillus however presents linear peptides on its cell wall. Small autoinducing peptides of a downstream signaling system could be released by the cleavage of these membrane-anchored cell surface proteins. This involves including a protease cleavage site between the membrane anchored domain and the autoinducing peptide. Proteases released from the Circaria parasite would target this site and hence we can detect them as our system becomes activated. | Bacillus uniquely utilizes a quorum sensing system based on using small peptides as autoinducers. Many other bacteria including gram negative bacteria use a QSS based on circular or post-translationally modified peptide display systems; this introduced problems with our desire to re-engineer the small peptide fragment. Bacillus however presents linear peptides on its cell wall. Small autoinducing peptides of a downstream signaling system could be released by the cleavage of these membrane-anchored cell surface proteins. This involves including a protease cleavage site between the membrane anchored domain and the autoinducing peptide. Proteases released from the Circaria parasite would target this site and hence we can detect them as our system becomes activated. | ||
Protease detector peptides (small linear quorum sensing peptide attached to protease recognition sequence) may also get trapped between the cell wall and cell membrane. To avoid this we looked into mechanisms of transport and attachment of these peptides to the outside of cell wall. An alternative was secrete detector peptides into the medium as B.subtilis is very good at secreting proteins. The lytC cell wall delivery and anchoring system was chosen to satisfy our system of peptide presentation. Specifically, our protease cleavage site linker peptide plus autoinducing peptide were attached to the cell wall binding domain of an endogenous (to Bacillus) LytC surface protein. | Protease detector peptides (small linear quorum sensing peptide attached to protease recognition sequence) may also get trapped between the cell wall and cell membrane. To avoid this we looked into mechanisms of transport and attachment of these peptides to the outside of cell wall. An alternative was secrete detector peptides into the medium as B.subtilis is very good at secreting proteins. The lytC cell wall delivery and anchoring system was chosen to satisfy our system of peptide presentation. Specifically, our protease cleavage site linker peptide plus autoinducing peptide were attached to the cell wall binding domain of an endogenous (to Bacillus) LytC surface protein. | ||
| - | + | N.B. An additional option considered was to attach the small protease detector peptides on to pili (fimbriae) that are normally presented on the bacterial cell. This has been done routinely on E.coli fimbriae at non-conserved regions and outer membrane proteins to present various antigens [[1]]. There is also a good review of a variety of export patways that have been used [[2]] in gram negative bacteria. There is a lot work done on gram positive bacteria, although there is a detailed and very accessible review [[3]] of current understanding of G+ve cell surface. | |
| Line 65: | Line 66: | ||
|- | |- | ||
| - | ! Issues:!! Solution: | + | ! scope="col" width="225" | Issues:!! scope="col" width="225" | Solution: |
|- | |- | ||
| | | | ||
| Line 83: | Line 84: | ||
|- | |- | ||
| - | ! Required components !! Features of ''B. subtilis' | + | ! scope="col" width="225" | Required components !! scope="col" width="225" | Features of ''B. subtilis' |
|- | |- | ||
| | | | ||
| Line 94: | Line 95: | ||
'''Notes:''' | '''Notes:''' | ||
| + | |||
Bacillus is a naturally occurring soil bacterium that presents no pathogenicity to humans. We aim to introduce our new DNA into existing essential genes. The disruption of these genes render the bacteria unlikely to exist outside of the lab or eventual product environment. | Bacillus is a naturally occurring soil bacterium that presents no pathogenicity to humans. We aim to introduce our new DNA into existing essential genes. The disruption of these genes render the bacteria unlikely to exist outside of the lab or eventual product environment. | ||
| Line 103: | Line 105: | ||
|- | |- | ||
| - | ! Required components !! Features of ''B. subtilis' | + | ! scope="col" width="225" | Required components !! scope="col" width="225" | Features of ''B. subtilis' |
|- | |- | ||
| | | | ||
| Line 114: | Line 116: | ||
'''Notes:''' | '''Notes:''' | ||
| + | |||
Luckily for us B.subtilis was the chassis of choice of the IC iGEM 2008 team. This means that there are now Bacillus friendly parts in the registry that we can use and also further characterize and contribute towards. | Luckily for us B.subtilis was the chassis of choice of the IC iGEM 2008 team. This means that there are now Bacillus friendly parts in the registry that we can use and also further characterize and contribute towards. | ||
| Line 120: | Line 123: | ||
|- | |- | ||
| - | ! Required components !! Features of ''B. subtilis' | + | ! scope="col" width="225" | Required components !! scope="col" width="225" | Features of ''B. subtilis' |
|- | |- | ||
| | | | ||
Revision as of 21:25, 24 October 2010
| Chassis |
|
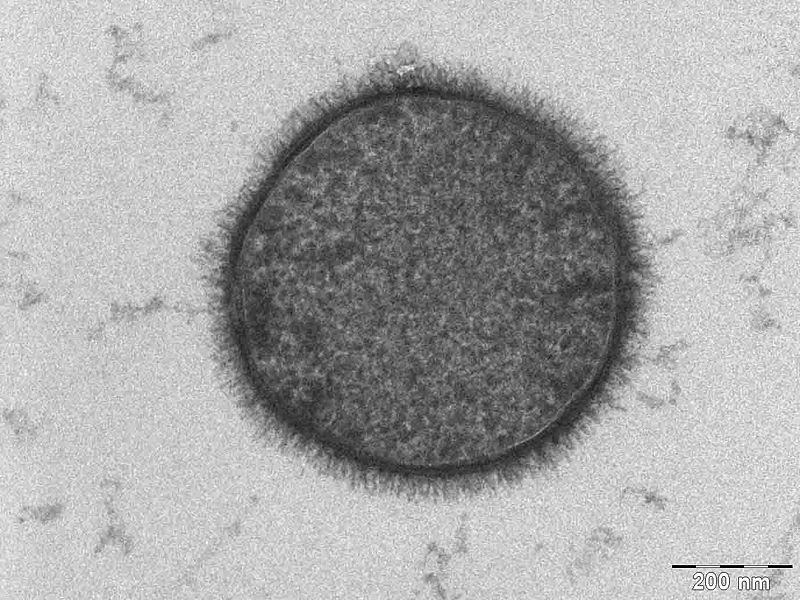
Our choice of chassis was B. subtilis. Here we list the reasons why B. subtilis serves as a more appropriate chassis option with regards to our project design and main project specifications. To re-iterate the pertinent project specifications:
We selected B. subtilis because it is a well characterized gram positive bacterium that is non pathogenic. It was convenient but also a coincidence that within our chosen chassis were features with which we could manipulate to use in our detection module. Below is a brief table summary of how Bacillus meets our system requirements and also some of the issues we encountered throughout the implementation process and how we overcame them. |
| Bacillus | Breakdown | ||||||||||||||||||||
|
Notes: Bacillus uniquely utilizes a quorum sensing system based on using small peptides as autoinducers. Many other bacteria including gram negative bacteria use a QSS based on circular or post-translationally modified peptide display systems; this introduced problems with our desire to re-engineer the small peptide fragment. Bacillus however presents linear peptides on its cell wall. Small autoinducing peptides of a downstream signaling system could be released by the cleavage of these membrane-anchored cell surface proteins. This involves including a protease cleavage site between the membrane anchored domain and the autoinducing peptide. Proteases released from the Circaria parasite would target this site and hence we can detect them as our system becomes activated. Protease detector peptides (small linear quorum sensing peptide attached to protease recognition sequence) may also get trapped between the cell wall and cell membrane. To avoid this we looked into mechanisms of transport and attachment of these peptides to the outside of cell wall. An alternative was secrete detector peptides into the medium as B.subtilis is very good at secreting proteins. The lytC cell wall delivery and anchoring system was chosen to satisfy our system of peptide presentation. Specifically, our protease cleavage site linker peptide plus autoinducing peptide were attached to the cell wall binding domain of an endogenous (to Bacillus) LytC surface protein. N.B. An additional option considered was to attach the small protease detector peptides on to pili (fimbriae) that are normally presented on the bacterial cell. This has been done routinely on E.coli fimbriae at non-conserved regions and outer membrane proteins to present various antigens 1. There is also a good review of a variety of export patways that have been used 2 in gram negative bacteria. There is a lot work done on gram positive bacteria, although there is a detailed and very accessible review 3 of current understanding of G+ve cell surface.
Safety
Notes: Bacillus is a naturally occurring soil bacterium that presents no pathogenicity to humans. We aim to introduce our new DNA into existing essential genes. The disruption of these genes render the bacteria unlikely to exist outside of the lab or eventual product environment.
Notes: Luckily for us B.subtilis was the chassis of choice of the IC iGEM 2008 team. This means that there are now Bacillus friendly parts in the registry that we can use and also further characterize and contribute towards.
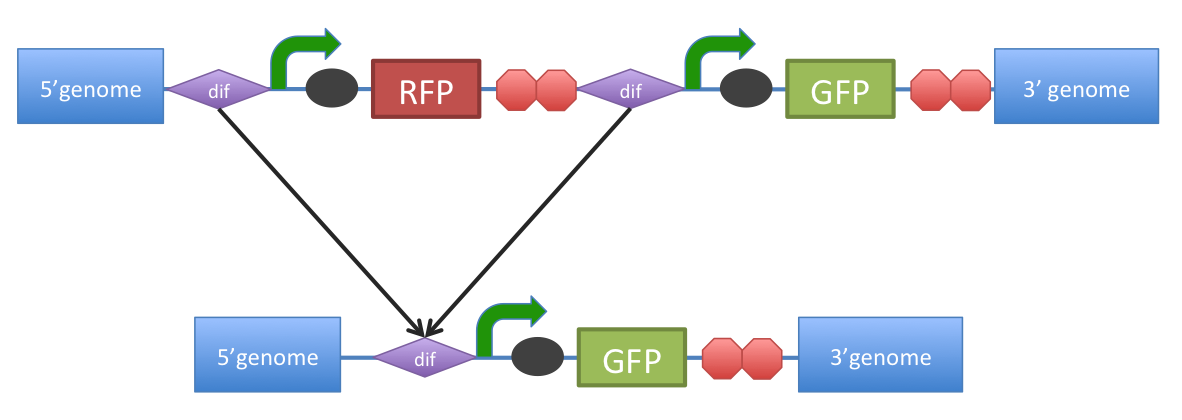
B. subtilis dif: sequence-specific recombinase recognition siteIn cells with circular chromosomes, recombinatorial repair and homologous recombination can generate multimeric chromosomes 1. Dif sites are part of a system to ensure that multimeric chromosomes can be separated to monomers, which is required for proper sharing of genetic material between daughter cells. In B. subtilis the tyrosine family recombinases such as RipX and CodV mediate the separation at 28-bp sequence Bs dif 1. The site-specific recombinases are able to recognize two directly repeated dif sites and excise the fragment flanked by the two sites 2.
Figure 1. Removal of a specific gene from a genome integrated construct.The site-specific recombinases, endogenous to ‘’B. subtilis’’ strains are able to recognise Bs’’dif’’ sites and recombine out the strand of DNA directly flanked by the two sites. Recombination leaves a single dif site. The construct was previously engineered to homologously recobine into the genome of B. subtilis. Integration sequences such as amyE <bbpart>BBa_K143001</bbpart>, <bbpart>BBa_K143002</bbpart> can be used to achieve this.
Sequence and Features <partinfo>BBa_K316002 SequenceAndFeatures</partinfo>
1) Sciochetti SA, Piggot PJ, and Blakely GW. Identification and characterization of the dif Site from Bacillus subtilis. J Bacteriol 2001 Feb; 183(3) 1058-68. doi:10.1128/JB.183.3.1058-1068.2001 pmid:11208805. PubMed HubMed 2) Bloor AE and Cranenburgh RM. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl Environ Microbiol 2006 Apr; 72(4) 2520-5. doi:10.1128/AEM.72.4.2520-2525.2006 pmid:16597952.
E.coli was considered as a possible option. Despite the gram negative outer membrane, there are strains that have been made more permeable through knockout of Lipid A biosynthesis in lipopolysaccharides [4] There are also proteins that can disrupt membranes when they are inserted in them, thus making them more permeable. These chages in permeability are likely due to transient ruptures of outer membrane and so unlikely to make a very responsive or robust detecton organism [4]. |
| References |
|
Surplus information on B. Subtilis chassis [http://www.subtiwiki.uni-goettingen.de/wiki/index.php/Main_Page Subti-Wiki] [http://mic.sgmjournals.org/cgi/content/full/146/12/3025 1] [http://www.microbialcellfactories.com/content/5/1/22 2] [http://www3.interscience.wiley.com/cgi-bin/fulltext/118603263/PDFSTART 3] [http://aac.asm.org/cgi/content/full/43/6/1459?view=long&pmid=10348770 4] |
 "
"