Igem2010/Main/synthetic miR Kit/October
From 2010.igem.org
(Difference between revisions)
Laura Nadine (Talk | contribs) |
|||
| Line 2: | Line 2: | ||
{{:Team:Heidelberg/Pagetop|note_mirna_kit}} | {{:Team:Heidelberg/Pagetop|note_mirna_kit}} | ||
= 01/10/2010 = | = 01/10/2010 = | ||
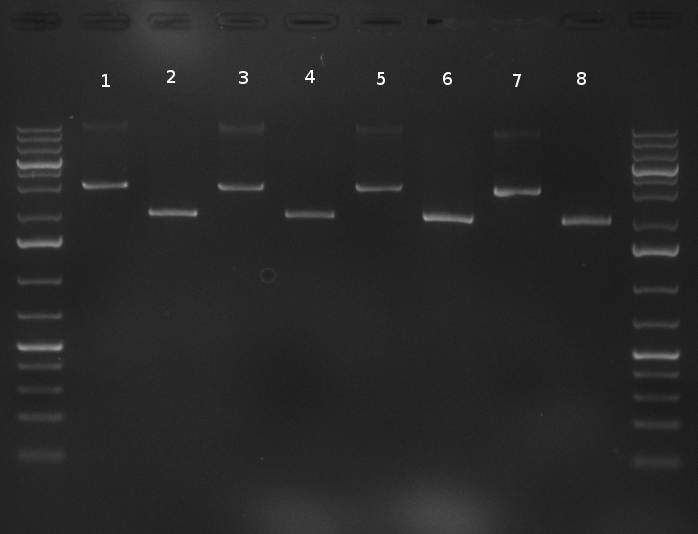
| - | [[Image:20101001_tetR_verdauEPanno.jpg|thumb| | + | [[Image:20101001_tetR_verdauEPanno.jpg|thumb|350px|right|Gel 101001-1. Digestion of miniprep no. 8, 9, 14 and 16 with EcoRI and PstI. Odd lanes are undigested controls, even lanes are digested minis]] |
repressor construct: | repressor construct: | ||
| Line 10: | Line 10: | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
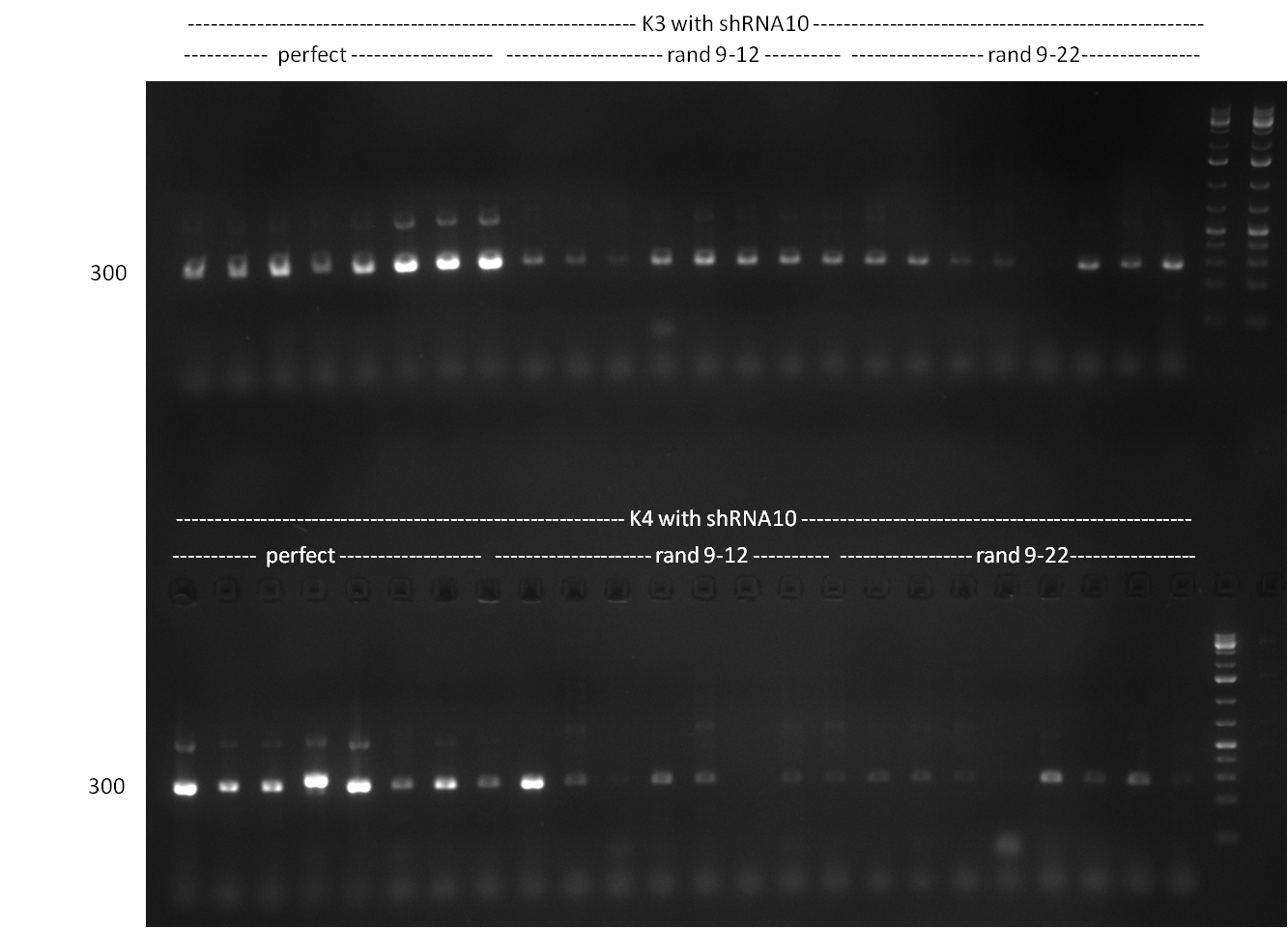
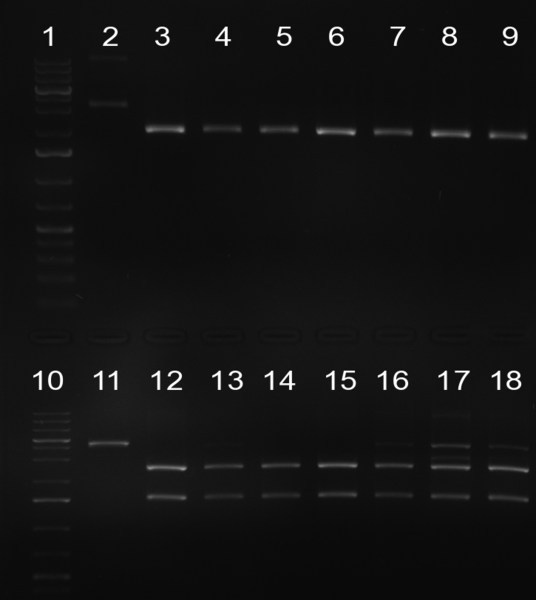
| - | [[Image:20101001_gel1_HD2010.png|thumb| | + | [[Image:20101001_gel1_HD2010.png|thumb|350 px|right|Screening 1: shRNA10 single binding sites]] |
| - | [[Image:20101001_gel2_HD2010.png|thumb| | + | [[Image:20101001_gel2_HD2010.png|thumb|350 px|right|Screening 2: shRNA10 single binding sites]] |
*Screening of the binding sites cloned into the constructs K3 and K4 containing shRNA10 in pSB1A3 on the previous day. 16 colonies were picked for each perfect, rand9-12 and rand9-22 binding site and colony-PCR was performed according to the colony-PCR standard protocol (Primers Luc2_bs_screen_seq and sh10_ape_xma_screen_rc). The results are shown on the gels Screening1 and Screening 2. Indicated are the construct numbers (either K3 or K4, which differ in the promoter driving the Luc2 gene) and the binding site property (perfect, 9-12 randomized or 9-22 randomized). Almost all clones show a positive band, although only some have a really bright band at the size of ~ 250 bp. LB cultures for 10 clones for each constructs K3 and K4 were inocculated. | *Screening of the binding sites cloned into the constructs K3 and K4 containing shRNA10 in pSB1A3 on the previous day. 16 colonies were picked for each perfect, rand9-12 and rand9-22 binding site and colony-PCR was performed according to the colony-PCR standard protocol (Primers Luc2_bs_screen_seq and sh10_ape_xma_screen_rc). The results are shown on the gels Screening1 and Screening 2. Indicated are the construct numbers (either K3 or K4, which differ in the promoter driving the Luc2 gene) and the binding site property (perfect, 9-12 randomized or 9-22 randomized). Almost all clones show a positive band, although only some have a really bright band at the size of ~ 250 bp. LB cultures for 10 clones for each constructs K3 and K4 were inocculated. | ||
| Line 18: | Line 18: | ||
= 02/10/2010 = | = 02/10/2010 = | ||
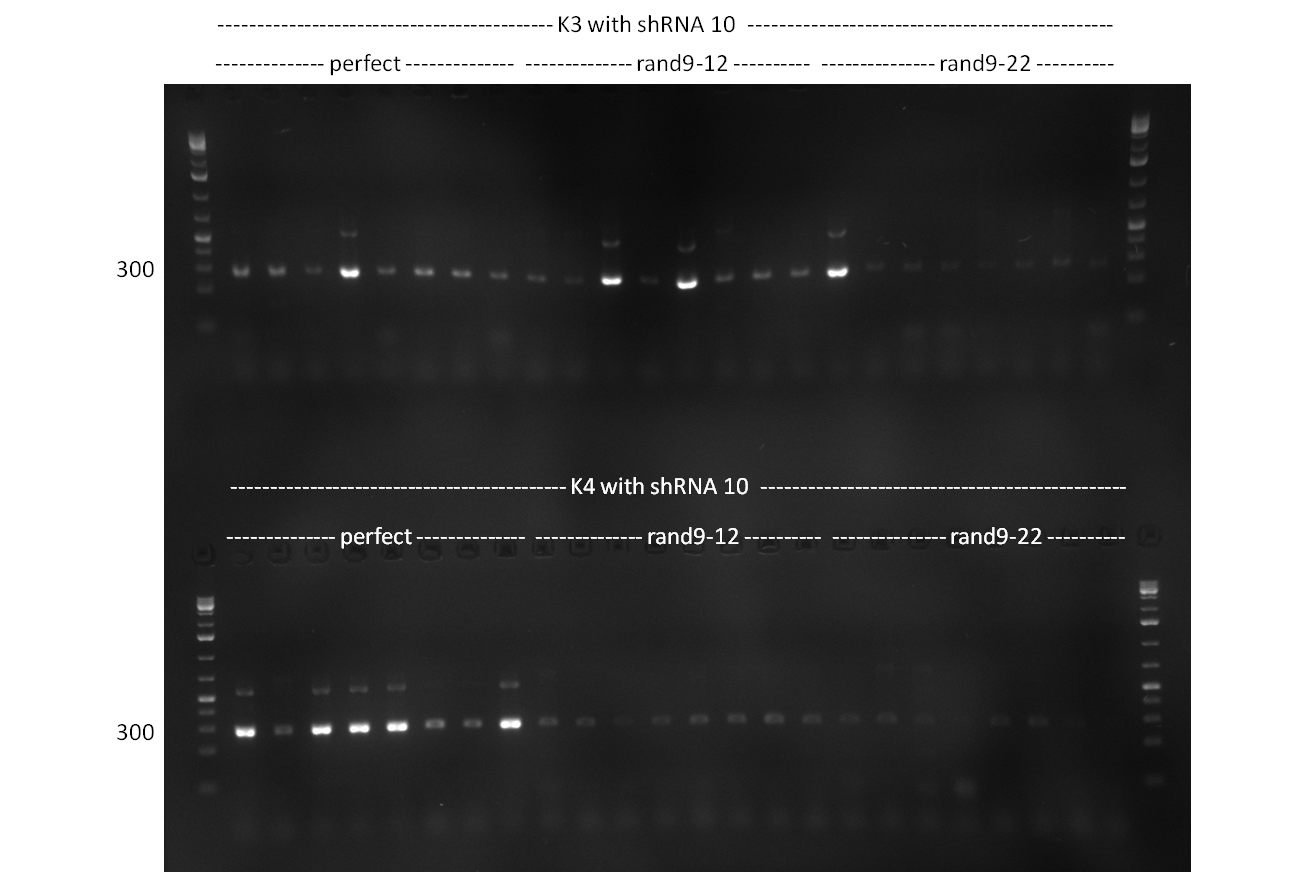
| - | [[Image:20101002 T1 T2 colonypcr.png|thumb| | + | [[Image:20101002 T1 T2 colonypcr.png|thumb|350px|right|Gel 101002-1. Colony PCR. All lanes with right amplified fragment except on lane 3, 10, 11, 15 and 23. Lane 1 & 14: 1kb Plus Ladder (Invitrogen)]] |
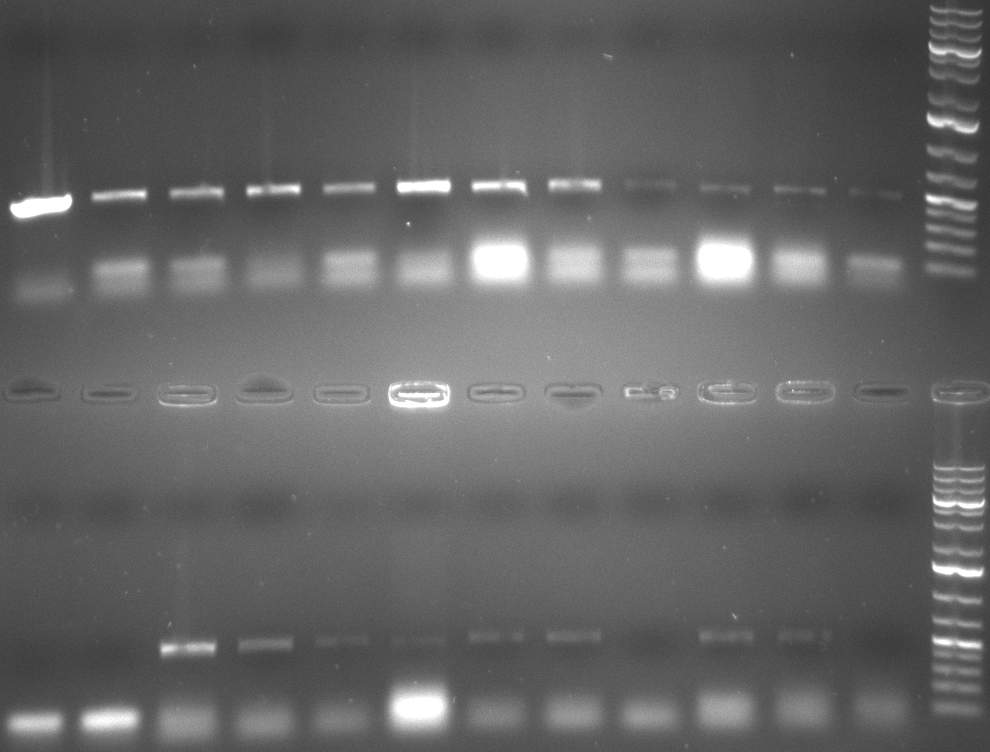
| - | [[Image:20101002 T1 T2 testdigest.png|thumb| | + | [[Image:20101002 T1 T2 testdigest.png|thumb|350px|right|Gel 101002-2. Test digestions of repressor construct. Upper pattern: EcoRI & PstI. Lower pattern: BamHI. All bands revealing positive results except in the first column (negative control from lane 3 of gel 101002-1). Lane 1 and Lane 10: 1kb Plus Ladder (Invitrogen). Lane 16, 17, 18: incomplete digestion ]] |
* Miniprep of the LB cultures (constructs K3 and K4 with shRNA10 and the corresponding binding sites) were done and used for transfection of Hek-293 cells | * Miniprep of the LB cultures (constructs K3 and K4 with shRNA10 and the corresponding binding sites) were done and used for transfection of Hek-293 cells | ||
| Line 95: | Line 95: | ||
= 10/10/2010 = | = 10/10/2010 = | ||
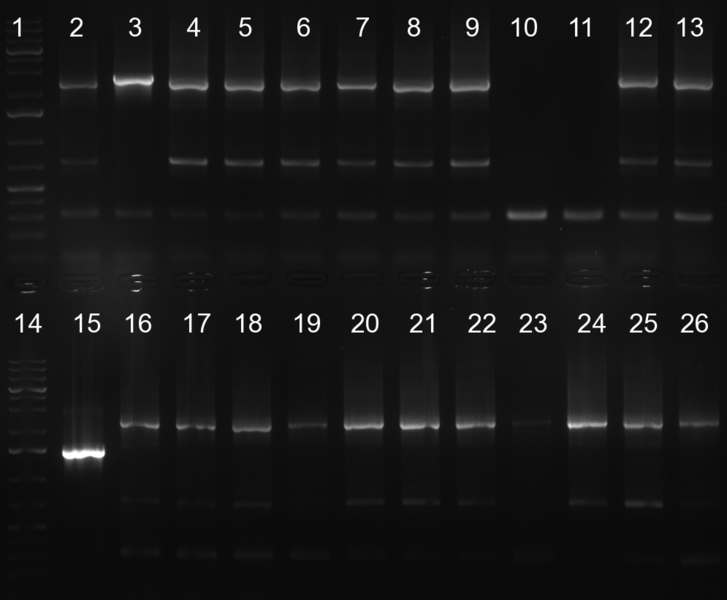
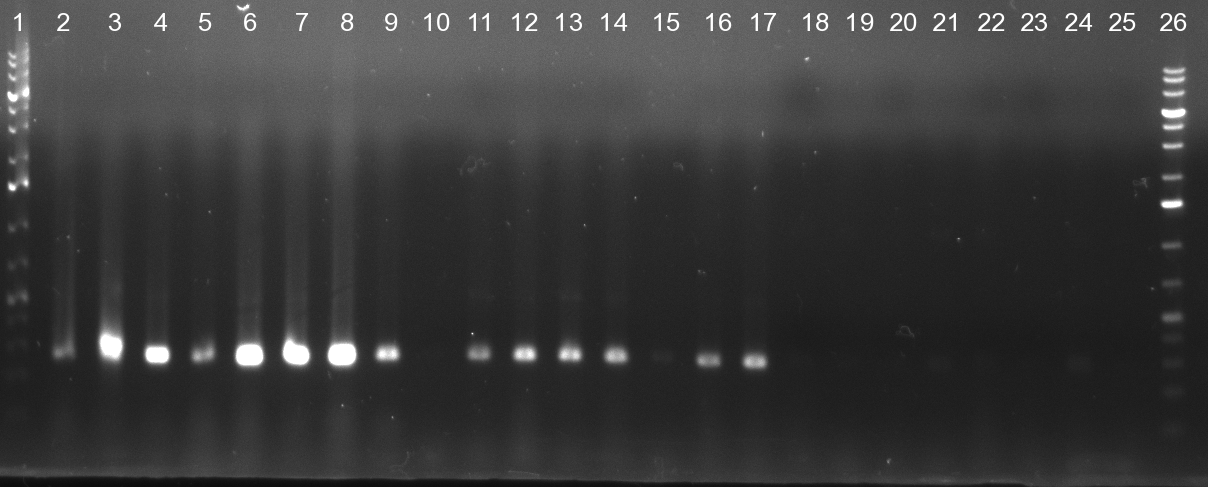
| - | [[Image:20101010 colonypcr shhAAT p 12 22 bs LOLA.png|thumb| | + | [[Image:20101010 colonypcr shhAAT p 12 22 bs LOLA.png|thumb|350px|right|Gel 101010-1. Colony PCR for construct containing shhAAT and binding sites. Expected is an amplified fragment a bit smaller than 300 bp which can be seen on lane 3, 4, 6, 7, 8 (perfect binding sites), 12, 13, 14, 16 and 17 (randomized binding sites). Lane 1 and Lane 26: 1kb Plus Ladder (Invitrogen)]] |
* colony pcr reveals some positive clones (see gel 101010-1) from [[Igem2010/Main/synthetic miR Kit/October#09/10/2010 | previous day]] | * colony pcr reveals some positive clones (see gel 101010-1) from [[Igem2010/Main/synthetic miR Kit/October#09/10/2010 | previous day]] | ||
** inoculation of 5 ml LB Amp cultures for mini-prep over night | ** inoculation of 5 ml LB Amp cultures for mini-prep over night | ||
| Line 140: | Line 140: | ||
=15/10/2010= | =15/10/2010= | ||
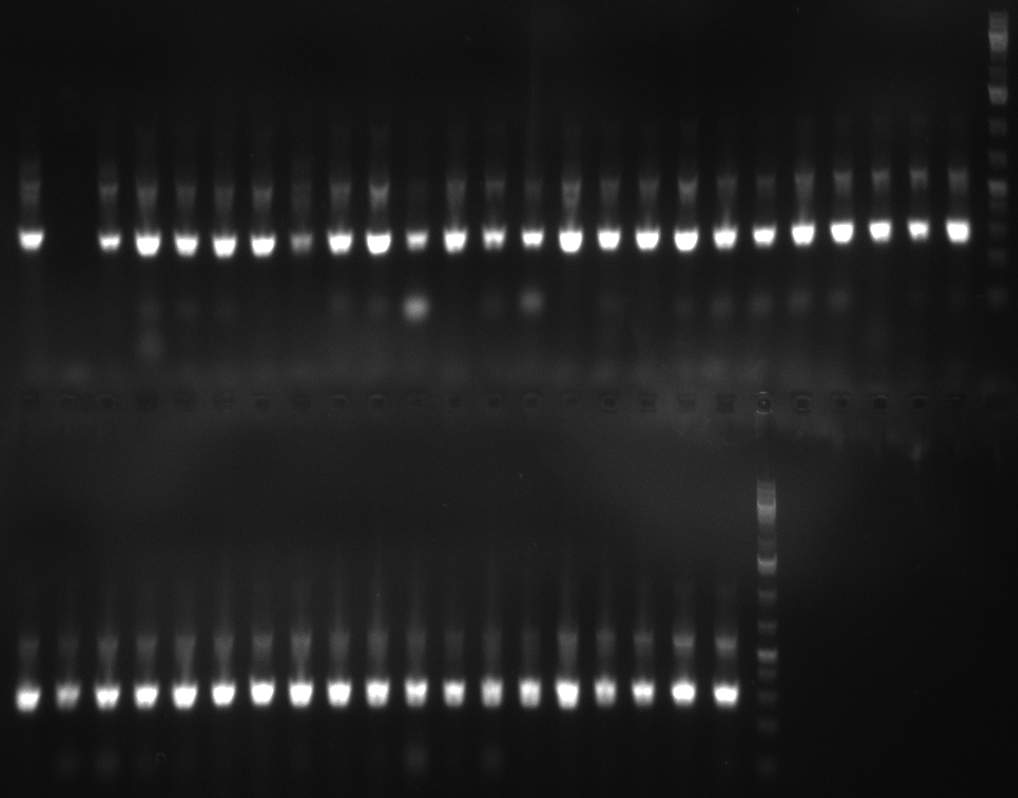
| - | [[Image:20101014_colonypcr_M1_M11anno.jpg|thumb| | + | [[Image:20101014_colonypcr_M1_M11anno.jpg|thumb|350px| Screening of M1 to M11 constructs, 4 clones were picked from each plate. 300bp band marks positive clones]] |
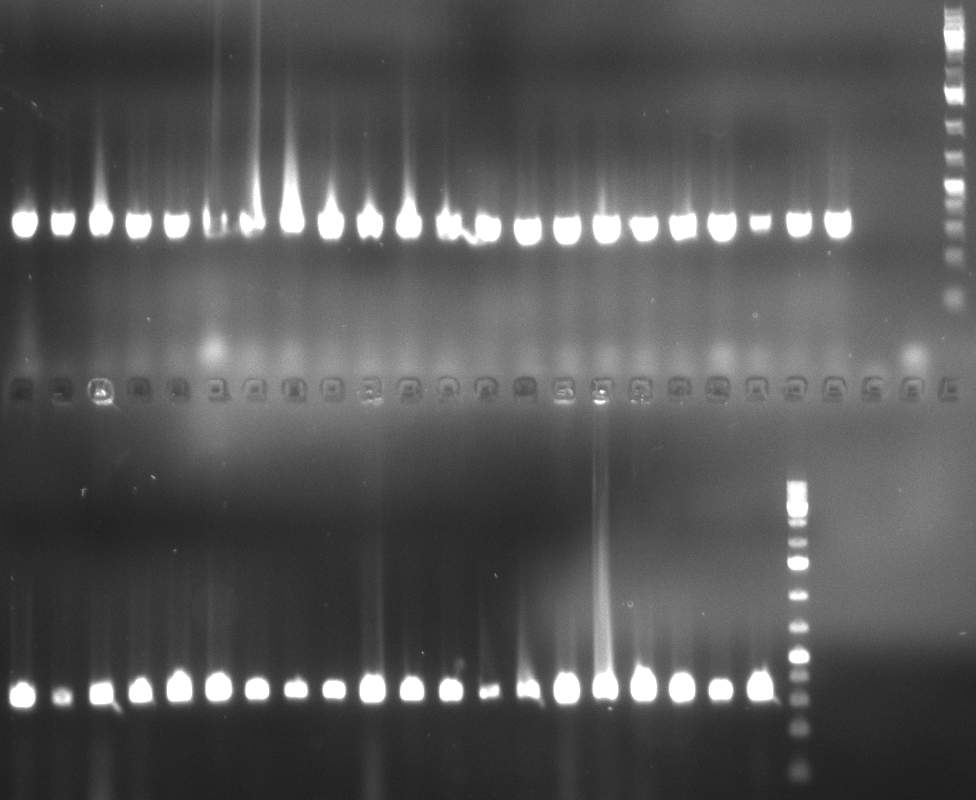
| - | [[Image:20101014_colonypcr_M12_M22anno.jpg|thumb| | + | [[Image:20101014_colonypcr_M12_M22anno.jpg|thumb|350px| Screening of M12 to M22 constructs, 4 clones were picked from each plate. 300bp band marks positive clones]] |
| - | [[Image:20101014_colonypcr_mimeasure122BS_p_12_22anno.jpg|thumb| | + | [[Image:20101014_colonypcr_mimeasure122BS_p_12_22anno.jpg|thumb|350px| Screening of measurement standard with perfect, 9-12 and 9-22 randomized binding sites. constructs, 8 clones were picked from each plate. 300bp band marks positive clones]] |
* screening of plates M1 to M11 of the transformation with the construct pBSU6_SV40_Luc2 with 11 different binding sites for sh hAAT. Four clones from each plate were screened by colony pcr, positives give a band at 300bp. Primers Luc2_bs_screen_seq_fw and reverse primerse are the reverse binding site oligos for the binding sites. | * screening of plates M1 to M11 of the transformation with the construct pBSU6_SV40_Luc2 with 11 different binding sites for sh hAAT. Four clones from each plate were screened by colony pcr, positives give a band at 300bp. Primers Luc2_bs_screen_seq_fw and reverse primerse are the reverse binding site oligos for the binding sites. | ||
Revision as of 11:49, 25 October 2010

|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"