Team:Northwestern/Project/Modeling
From 2010.igem.org
(Difference between revisions)
Timsterxox (Talk | contribs) |
(→Modeling) |
||
| Line 154: | Line 154: | ||
=='''Modeling'''== | =='''Modeling'''== | ||
| + | |||
| + | Status: Under Development | ||
| + | |||
| + | The purpose of Modeling was to characterize expression vectors in terms of expression level and time-delay, all activated by a diffused inducer. | ||
| + | |||
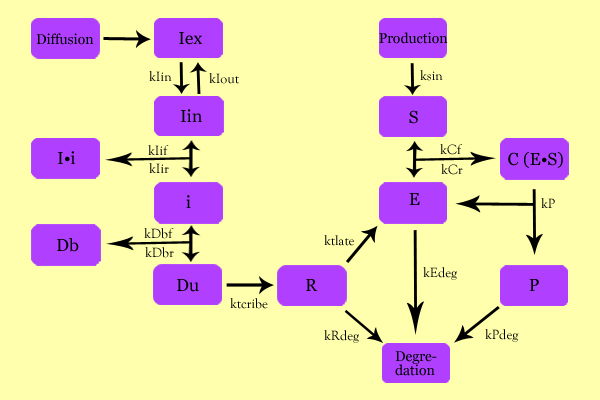
Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: | Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: | ||
[[Image:SuperModeling.jpg|500px|center]] | [[Image:SuperModeling.jpg|500px|center]] | ||
| - | |||
| - | Matlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion. | + | Variables: |
| + | Iex: External Inducer, determined by diffusion through Fick's law (IPTG in our experiment) | ||
| + | Iin: Internal Inducer (IPTG) | ||
| + | Ii: Inducer bound to Repressor (IPTG bound to lacI) | ||
| + | i: Repressor (lacI) | ||
| + | Db: Repressor-bound DNA (lacI-bound DNA(CHS3) region in plasmid) | ||
| + | Dunb: transcribe-able or Repressor-unbound DNA (lacI-unbound DNA(CHS3)) | ||
| + | Re: mRNA for Enzyme (CHS3 mRNA) | ||
| + | E: Enzyme (CHS3) | ||
| + | S: Substrate (N-Acetyl Glucosamine) | ||
| + | C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) | ||
| + | P: Protein Product (Chitin or (NAG)n+1) | ||
| + | |||
| + | Matlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion and initiate the process. | ||
The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. | The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. | ||
This model was fitted with empirical data using cp-lacpi-gfp to estimate the effect of varying cp, lacpi, and rbs on enzyme and final product production. | This model was fitted with empirical data using cp-lacpi-gfp to estimate the effect of varying cp, lacpi, and rbs on enzyme and final product production. | ||
| + | |||
| + | References: | ||
| + | A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures | ||
| + | William E. Bentley, Dhinakar S. Kompala | ||
| + | Article first published online: 18 FEB 2004 | ||
| + | |||
| + | DOI: 10.1002/bit.260330108 | ||
| + | http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf | ||
| + | |||
| + | Mathematical modeling of induced foreign protein production by recombinant bacteria | ||
| + | Jongdae Lee, W. Fred Ramirez | ||
| + | Article first published online: 19 FEB 2004 | ||
| + | |||
| + | DOI: 10.1002/bit.260390608 | ||
| + | http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf | ||
|} | |} | ||
Revision as of 23:15, 23 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
 "
"