Team:Lethbridge/Modeling
From 2010.igem.org
David.franz (Talk | contribs) (Putting up Jeff's homology blurb) |
David.franz (Talk | contribs) |
||
| Line 123: | Line 123: | ||
=<font color="white">Homology Modeling= | =<font color="white">Homology Modeling= | ||
| + | [[Image:homology1.png|x350px|right|text-top]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
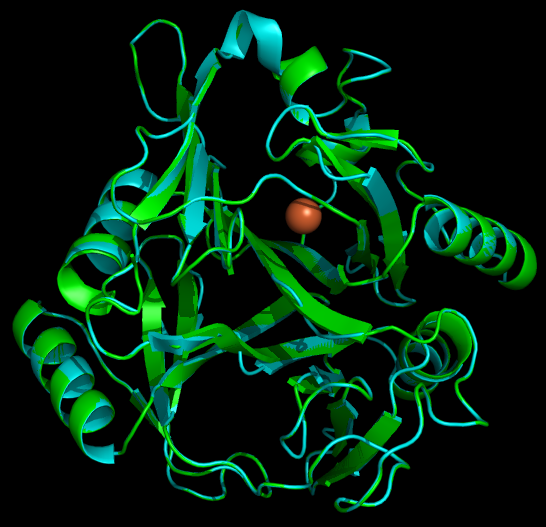
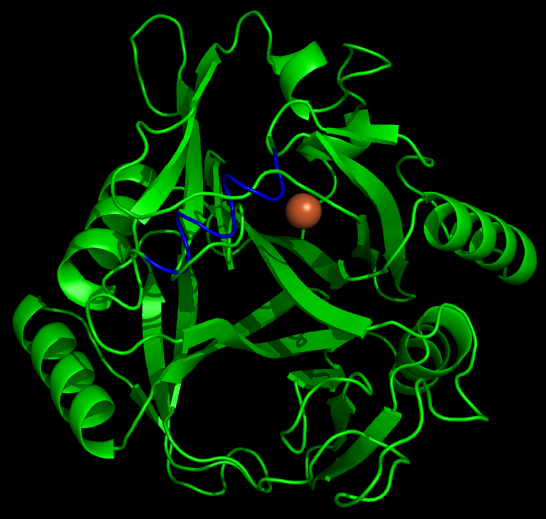
Another aspect of our project is working on the localization of catechol-2,3,-dioxygenase (and other proteins) into the interior of microcompartments. To target the protein into the microcompartment (Lumazine modified to have an even more charged interior) requires the fusion of a polyarginine tail to either the C or N terminus of the protein. | Another aspect of our project is working on the localization of catechol-2,3,-dioxygenase (and other proteins) into the interior of microcompartments. To target the protein into the microcompartment (Lumazine modified to have an even more charged interior) requires the fusion of a polyarginine tail to either the C or N terminus of the protein. | ||
| Line 128: | Line 135: | ||
==<font color="white">The Method and Results== | ==<font color="white">The Method and Results== | ||
| - | + | ||
| - | [[Image:homology2.png| | + | [[Image:homology2.png|x350px|right|text-top]] |
| + | |||
To model the XylE structure, the sequence for XylE from Pseudomonas putida (NCBI accession number NP_542866) was aligned with the primary sequence from the crystal structure of XylE from the same organism (pdb: 1MPY; several differences in amino acid sequence were observed) using CLUSTALW (1). Based on this sequence alignment, a homology model was generated using the alignment mode in SWISSMODEL (2-4). To model the placement of an N-terminal arginine tag, the tag was manually added to the N-terminus of the model. Energy minimization was carried out in SWISS-PDB viewer in vacuo utilizing a GROMOS96 energy minimization (2). The resulting pdb file was visualized and manipulated using PYMOL, images were taken using the same software (5). | To model the XylE structure, the sequence for XylE from Pseudomonas putida (NCBI accession number NP_542866) was aligned with the primary sequence from the crystal structure of XylE from the same organism (pdb: 1MPY; several differences in amino acid sequence were observed) using CLUSTALW (1). Based on this sequence alignment, a homology model was generated using the alignment mode in SWISSMODEL (2-4). To model the placement of an N-terminal arginine tag, the tag was manually added to the N-terminus of the model. Energy minimization was carried out in SWISS-PDB viewer in vacuo utilizing a GROMOS96 energy minimization (2). The resulting pdb file was visualized and manipulated using PYMOL, images were taken using the same software (5). | ||
 "
"