Team:TU Munich/Project
From 2010.igem.org
(→Vision) |
(→Vision) |
||
| Line 22: | Line 22: | ||
*Designing a synthetic version of a RNA-sensitive riboswitch, which is based on the principle of shifting between a state of antitermination and termination as observed for attenuator operons in nature. [[Image:Minilink.gif | link]] | *Designing a synthetic version of a RNA-sensitive riboswitch, which is based on the principle of shifting between a state of antitermination and termination as observed for attenuator operons in nature. [[Image:Minilink.gif | link]] | ||
*Coupling this principle to the phenomenon of tiny abortive RNA´s, which cause antitermination in T7-termintors. [[Image:Minilink.gif | link]] | *Coupling this principle to the phenomenon of tiny abortive RNA´s, which cause antitermination in T7-termintors. [[Image:Minilink.gif | link]] | ||
| - | + | <br> | |
To evaluate functionality of our molecular switches, we developed several [[in vivo and in vitro assays| in vivo and in vitro assays]] and relied on [[existing assays| existing assays]]. | To evaluate functionality of our molecular switches, we developed several [[in vivo and in vitro assays| in vivo and in vitro assays]] and relied on [[existing assays| existing assays]]. | ||
Thus we don´t want to develop additional biobricks, we rather want to establish a platform to realize the full opportunity of the biobrick system. This would enable people easily setting up sensor - reporter circuits AND interconnect them to complete biological chips... the way to real artificial cells and synthetic biology | Thus we don´t want to develop additional biobricks, we rather want to establish a platform to realize the full opportunity of the biobrick system. This would enable people easily setting up sensor - reporter circuits AND interconnect them to complete biological chips... the way to real artificial cells and synthetic biology | ||
Revision as of 13:08, 20 October 2010
|
||||||||
|
|
VisionUntil today, 13.628 biobrick sequences[1] have been submitted to partsregistry, thereof 102 reporter units, 12 signaling bricks and xx sensing parts.
Since there, people are trying to arrange these single biological building blocks in such a manner that allows producing special biotechnological products (metabolic engineering), developing biological sensory circuits (biosensors) and even giving microorganisms the ability to react on multiple environmental factors and serve both as disease indicator and drug. These examples and further promising ideas were implemented on previous iGEM-competitions.[2][3][4]
Our switches provide the opportunity to use one single switching mechanism and only vary the recognition unit, to built numerous "transistors" - meaning the same technical principle is used, but only the interconnection of cables (in our case signal rna and respective recognition site) has to be changed. .... To evaluate functionality of our molecular switches, we developed several in vivo and in vitro assays and relied on existing assays Perequesites
Reference[1] http://partsregistry.org/cgi/partsdb/Statistics.cgi
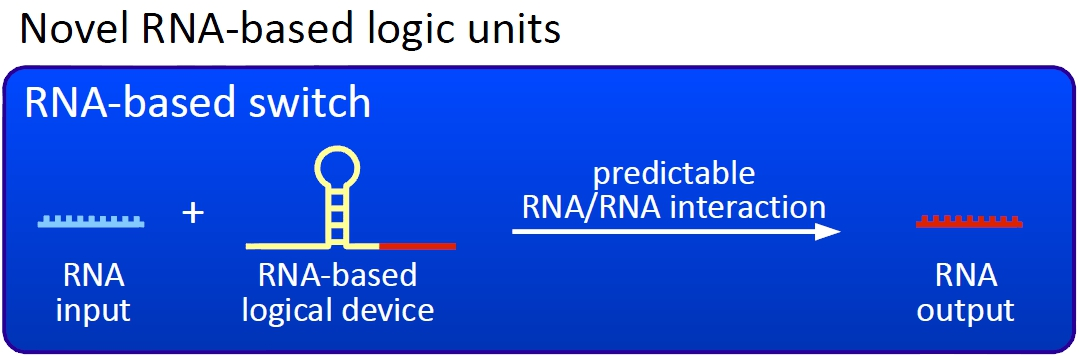
ConceptThe basic principle of our switches are short RNA sequences, the scientific idea shares similiarities with the principle of antitermination but also inherits a completely new way of RNA based transcription regulation. We used three-dimensional structure predictions and thermodynamic calculation to develop a set of switches - about 50 nucleotides - and signals - about 20 nucleotides. The switch forms a stem loop causing transcription termination, which can be resolved upon binding of a signal. On/off switching can therefore be easily controlled by signal avaiability and provides a new concept to control gene transcription.
Our switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences as transcriptional switches, that are calculated to be capable of stem loop formation. We plan to control the termination at these switches with a small RNA molecule (our RNA "signal") that is complementary to a part of the stem loop forming sequence. This small functional RNA inhibits the stem loop formation by complementary base-pairing and hence avoids transcription termination. The signal is composed of two parts: While the first part provides specifity (recognition site), the second part causing stem loop disintegration (functional core) can in principle be the same for all bioLOGICS. Therefore variation of the first part allows the construction of an endless number of switches. The functional core causing stem loop disintegration is based on a working system (see attenuation) established by nature. Different stem loops were tested in this effort: Regulatory parts from the E. coli trp-operon, his-operon and one based on previous iGEM-work.
Network constructionDesigning complex biological networks based on either traditional protein engineering or our new bioLOGICS is still a complex task. We developed a software which allows the fast construction of a bioLOGICS based networks. Work ProgressEvery network starts with a basic unit. While our declared aim is to enable networks allowing fine-tuning of gene expression beyond the regular on/off, exploring such an on/off switch/signal pair is the first step towards a functional network. We constructed several units and tested their efficiency, robustness and reproducibility in vivo, in vitro and in silico. Furthermore we developed a software which allows easy constructions of networks delivering a ready network. Conclusive elaboration of a few first RNA-based logic units is the major contribution of our iGEM team. Results |
|||||||
 "
"