Team:BCCS-Bristol/Wetlab/Part Design/BioBricks/PyeaR+LacZ
From 2010.igem.org
(→LacZ BioBrick) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:BCCS-Bristol/Header}} | {{:Team:BCCS-Bristol/Header}} | ||
| - | + | <center> • [[:Team:BCCS-Bristol/Wetlab/Part_Design/BioBricks/PyeaR|BBa_K381001]] • [[:Team:BCCS-Bristol/Wetlab/Part_Design/Components/Promoters|Promoter]] • [[:Team:BCCS-Bristol/Wetlab/Part_Design/Components/Reporters|Reporters]] | |
| + | • [[:Team:BCCS-Bristol/Wetlab/Part_Design/BioBricks/PyeaR%2BLacZ|PyeaR + LacZ construct]] • [[:Team:BCCS-Bristol/Wetlab/Part_Design/Components/LacZ|LacZ]] • </center> | ||
| + | = PyeaR + LacZ Construct = | ||
| - | The original plan was to use the [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part to characterise the PyeaR promoter, through a β-galactosidase assay to determine the activity of the | + | ==Motivation== |
| + | {{:Team:BCCS-Bristol/newtoc}} | ||
| + | The original plan was to use the [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part to characterise the PyeaR promoter, through a β-galactosidase assay to determine the activity of the PyeaR part. Having struggled to get a workable result through Edinburgh's BioBrick, we decided to set about designing our own. | ||
| + | |||
| + | The plan would be to take the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] part and use this as a base for the new part containing a full functional copy of the LacZ part - for this part an ideal BioBrick in the registry already existed, containing a good Ribosomal Binding Site (RBS) plus the full length LacZ sequence: [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]. Combining the two existing biobricks into one should have been a simple procedure, though through error and plain bad luck, after to consecutive attempts no working product was forthcoming, and with just two weeks before the new term was due to start we abandoned the construction of the PyeaR + LacZ construct in favour of getting the best data we could from Edinburgh's [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part. | ||
| + | |||
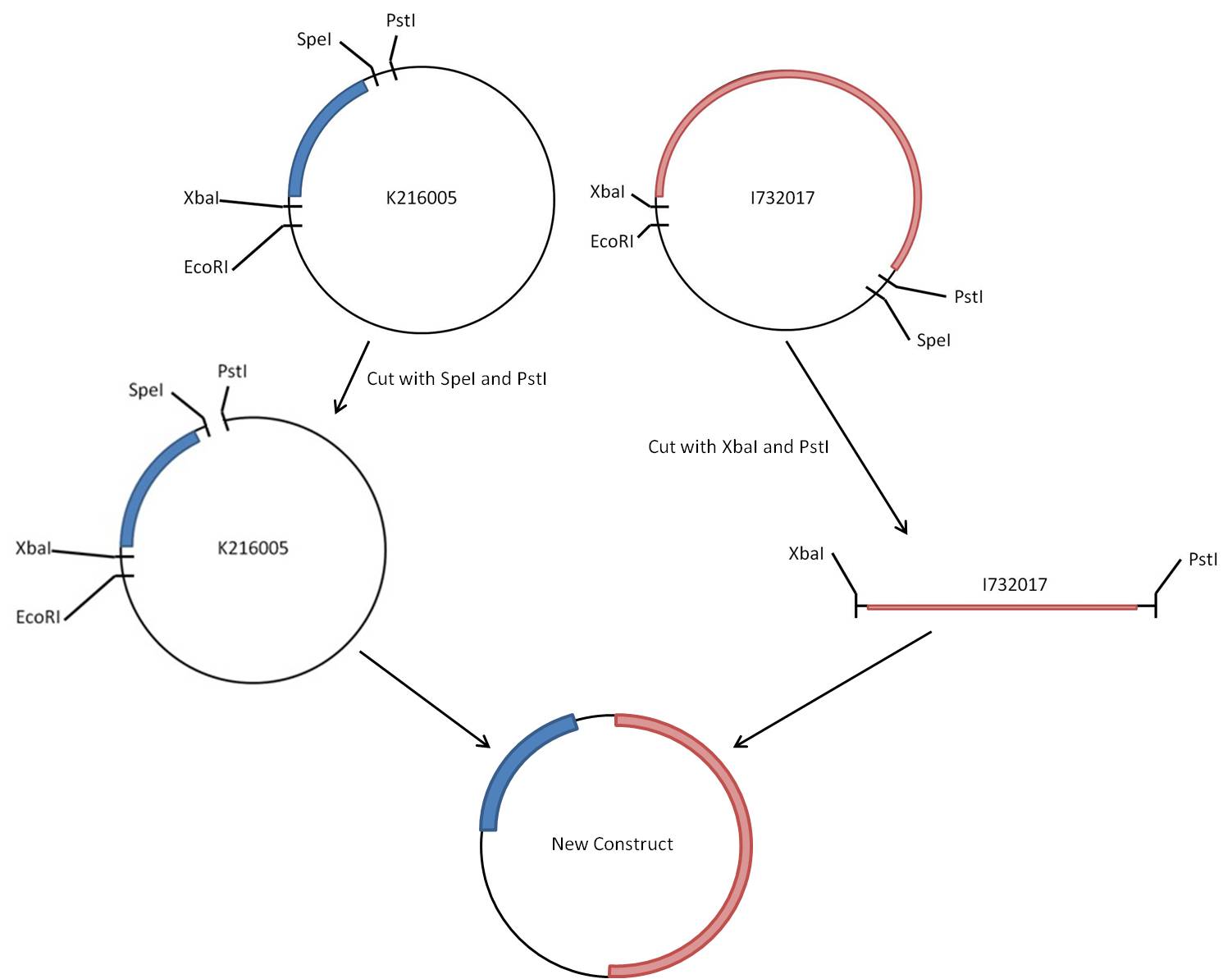
| + | The rest of this page is dedicated to the procedure followed in the attempt at creating the PyeaR + LacZ construct, and also points out some of the difficulties encountered along the way. The first figure is a virtual map of the construction of the new BioBrick, containing the barest details of the work that went in. | ||
| + | |||
| + | [[Image:BCCS VirtualDigestMap LacZConstuct.jpg|frameless|center|upright=4|PyeaR+LacZConstructMap]] | ||
| + | |||
| + | The steps to achieve the new construct are as follows, assuming you have pure concentrated stocks of both original biobricks ([http://partsregistry.org/Part:BBa_K216005 BBa_K216005] & [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]): | ||
| + | |||
| + | |||
| + | ==Procedure== | ||
| + | |||
| + | '''Step one''' - double digest of both BioBricks. | ||
| + | |||
| + | *[http://partsregistry.org/Part:BBa_K216005 BBa_K216005] | ||
| + | **30.0μL Purified [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] Plasmid DNA | ||
| + | **0.5μL BSA 100x | ||
| + | **5.0μL NEBuffer 2 10x | ||
| + | **1.0μL PstI | ||
| + | **1.3μL SpeI | ||
| + | **12.2μL ddH<sub>2</sub>O | ||
| + | Total volume: 50.0μL | ||
| + | |||
| + | *[http://partsregistry.org/Part:BBa_I732017 BBa_I732017] | ||
| + | **30.0μL Purified [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] Plasmid DNA | ||
| + | **0.5μL BSA 100x | ||
| + | **5.0μL NEBuffer 3 10x | ||
| + | **1.0μL PstI | ||
| + | **1.3μL XbaI | ||
| + | **12.2μL ddH<sub>2</sub>O | ||
| + | Total volume: 50.0μL | ||
| + | |||
| + | Both were incubated at 37°C, [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] for 2 hours and [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] for 3 hours. After 2 hours, the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] digestion mix was removed from incubation. | ||
| + | |||
| + | '''Step 2''' - phosphatase treatment of [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] to prevent recircularisation. | ||
| + | |||
| + | *[http://partsregistry.org/Part:BBa_K216005 BBa_K216005] phosphatase treatment | ||
| + | **50.0μL [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] digestion mix | ||
| + | **7.0μL alkaline phosphatase buffer 10x | ||
| + | **7.0μL alkaline phosphatase | ||
| + | **6.0μL ddH<sub>2</sub>O | ||
| + | Total volume: 70.0μL | ||
| + | |||
| + | Placed back in 37°C incubation for 1 hour. | ||
| + | |||
| + | '''Step 3''' - Agarose gel | ||
| + | |||
| + | Added <sup>1</sup>/<sub>5</sub> total volume loading buffer to both the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] phosphatase mix (~14.0μL) and the [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] double digest mix (~10.0μL). Ran the gel at 100V for 2 hours with a 1kb DNA ladder for reference. | ||
| + | |||
| + | '''Step 4''' - Gel purification | ||
| + | |||
| + | Selection of appropriate bands on gel to purify using a gel purification kit. | ||
| + | |||
| + | '''Step 5''' - Ligation | ||
| + | |||
| + | *[http://partsregistry.org/Part:BBa_K216005 BBa_K216005]+[http://partsregistry.org/Part:BBa_I732017 BBa_I732017] ligation mix | ||
| + | **2.0μL purified cut [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] | ||
| + | **6.0μL purified cut [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] | ||
| + | **2.0μL 10x T4 ligase buffer | ||
| + | **1.0μL T4 ligase | ||
| + | **9.0μL ddH<sub>2</sub>O | ||
| + | |||
| + | *Control mix | ||
| + | **2.0μL purified cut [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] | ||
| + | **2.0μL 10x T4 ligase buffer | ||
| + | **1.0μL T4 ligase | ||
| + | **15.0μL ddH<sub>2</sub>O | ||
| + | |||
| + | Both ligations incubated on ice overnight. | ||
| + | |||
| + | '''Step 6''' - Checking ligations by agarose gel electrophoresis | ||
| + | |||
| + | Grow up ligated plasmid in a cloning strain of ''E.coli'', set up cultures and miniprep DNA to produce purified Plasmid DNA. Set up double digest of ligation mix as follows: | ||
| + | |||
| + | *Ligation mix double digest | ||
| + | **3.0μL purified ligated plasmid DNA | ||
| + | **2.0μL NEBuffer 2 10x | ||
| + | **0.2μL BSA 10X | ||
| + | **0.5μL SpeI | ||
| + | **0.5μL XbaI | ||
| + | **13.8μL ddH<sub>2</sub>O | ||
| + | Total volume: 20.0μL | ||
| + | |||
| + | Incubated for 2 hours at 37°C. Add <sup>1</sup>/<sub>5</sub> total volume loading buffer to the digest mix (~4.0μL). Ran agarose gel at 100V for 2 hours, including a 1kb DNA ladder for reference. | ||
| + | |||
| + | ==Notes on experiments== | ||
| + | |||
| + | The same procedure was used for both attempts at creating the construct. The first failed attempt is fairly simple to explain. During '''Step 4''' the wrong band was chosen from the 3 which presented after running digested [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] on the gel. This occurred due to confusion concerning the size of the [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] insert compared to its vector - as it turns out the insert is over 1000bp longer than the vector. | ||
| + | |||
| + | The reason behind the second failed attempt is a little harder to pin down. For all the tests we ran as the procedure went on, the ligation appeared to have worked correctly. We even went on to attempt a β-galactosidase assay with the new construct, and achieved some fairly high activity with the plasmid. However, this activity did not correlate with the test KNO<sub>3</sub> concentrations (see the page on β-galactosidase assays for the data collected during this test, along with all the other repeats run during the project). With this information, we decided to have the construct sequenced, a procedure we would have to have done in any case, and awaited the results. | ||
| + | |||
| + | After around an hour of confusion on receiving the sequencing data, we found the reason behind the odd results - the PyeaR sequence had disappeared from the construct! With no explanation as to how this may have happened, we quickly rethought our plans and returned to the [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part in order to get some usable characterisation data, and sadly abandoned our attempt to create a PyeaR+LacZ construct. | ||
Latest revision as of 18:56, 27 October 2010
iGEM 2010
PyeaR + LacZ Construct
Motivation
The original plan was to use the [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part to characterise the PyeaR promoter, through a β-galactosidase assay to determine the activity of the PyeaR part. Having struggled to get a workable result through Edinburgh's BioBrick, we decided to set about designing our own.The plan would be to take the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] part and use this as a base for the new part containing a full functional copy of the LacZ part - for this part an ideal BioBrick in the registry already existed, containing a good Ribosomal Binding Site (RBS) plus the full length LacZ sequence: [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]. Combining the two existing biobricks into one should have been a simple procedure, though through error and plain bad luck, after to consecutive attempts no working product was forthcoming, and with just two weeks before the new term was due to start we abandoned the construction of the PyeaR + LacZ construct in favour of getting the best data we could from Edinburgh's [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part.
The rest of this page is dedicated to the procedure followed in the attempt at creating the PyeaR + LacZ construct, and also points out some of the difficulties encountered along the way. The first figure is a virtual map of the construction of the new BioBrick, containing the barest details of the work that went in.
The steps to achieve the new construct are as follows, assuming you have pure concentrated stocks of both original biobricks ([http://partsregistry.org/Part:BBa_K216005 BBa_K216005] & [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]):
Procedure
Step one - double digest of both BioBricks.
- [http://partsregistry.org/Part:BBa_K216005 BBa_K216005]
- 30.0μL Purified [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] Plasmid DNA
- 0.5μL BSA 100x
- 5.0μL NEBuffer 2 10x
- 1.0μL PstI
- 1.3μL SpeI
- 12.2μL ddH2O
Total volume: 50.0μL
- [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]
- 30.0μL Purified [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] Plasmid DNA
- 0.5μL BSA 100x
- 5.0μL NEBuffer 3 10x
- 1.0μL PstI
- 1.3μL XbaI
- 12.2μL ddH2O
Total volume: 50.0μL
Both were incubated at 37°C, [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] for 2 hours and [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] for 3 hours. After 2 hours, the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] digestion mix was removed from incubation.
Step 2 - phosphatase treatment of [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] to prevent recircularisation.
- [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] phosphatase treatment
- 50.0μL [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] digestion mix
- 7.0μL alkaline phosphatase buffer 10x
- 7.0μL alkaline phosphatase
- 6.0μL ddH2O
Total volume: 70.0μL
Placed back in 37°C incubation for 1 hour.
Step 3 - Agarose gel
Added 1/5 total volume loading buffer to both the [http://partsregistry.org/Part:BBa_K216005 BBa_K216005] phosphatase mix (~14.0μL) and the [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] double digest mix (~10.0μL). Ran the gel at 100V for 2 hours with a 1kb DNA ladder for reference.
Step 4 - Gel purification
Selection of appropriate bands on gel to purify using a gel purification kit.
Step 5 - Ligation
- [http://partsregistry.org/Part:BBa_K216005 BBa_K216005]+[http://partsregistry.org/Part:BBa_I732017 BBa_I732017] ligation mix
- 2.0μL purified cut [http://partsregistry.org/Part:BBa_K216005 BBa_K216005]
- 6.0μL purified cut [http://partsregistry.org/Part:BBa_I732017 BBa_I732017]
- 2.0μL 10x T4 ligase buffer
- 1.0μL T4 ligase
- 9.0μL ddH2O
- Control mix
- 2.0μL purified cut [http://partsregistry.org/Part:BBa_K216005 BBa_K216005]
- 2.0μL 10x T4 ligase buffer
- 1.0μL T4 ligase
- 15.0μL ddH2O
Both ligations incubated on ice overnight.
Step 6 - Checking ligations by agarose gel electrophoresis
Grow up ligated plasmid in a cloning strain of E.coli, set up cultures and miniprep DNA to produce purified Plasmid DNA. Set up double digest of ligation mix as follows:
- Ligation mix double digest
- 3.0μL purified ligated plasmid DNA
- 2.0μL NEBuffer 2 10x
- 0.2μL BSA 10X
- 0.5μL SpeI
- 0.5μL XbaI
- 13.8μL ddH2O
Total volume: 20.0μL
Incubated for 2 hours at 37°C. Add 1/5 total volume loading buffer to the digest mix (~4.0μL). Ran agarose gel at 100V for 2 hours, including a 1kb DNA ladder for reference.
Notes on experiments
The same procedure was used for both attempts at creating the construct. The first failed attempt is fairly simple to explain. During Step 4 the wrong band was chosen from the 3 which presented after running digested [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] on the gel. This occurred due to confusion concerning the size of the [http://partsregistry.org/Part:BBa_I732017 BBa_I732017] insert compared to its vector - as it turns out the insert is over 1000bp longer than the vector.
The reason behind the second failed attempt is a little harder to pin down. For all the tests we ran as the procedure went on, the ligation appeared to have worked correctly. We even went on to attempt a β-galactosidase assay with the new construct, and achieved some fairly high activity with the plasmid. However, this activity did not correlate with the test KNO3 concentrations (see the page on β-galactosidase assays for the data collected during this test, along with all the other repeats run during the project). With this information, we decided to have the construct sequenced, a procedure we would have to have done in any case, and awaited the results.
After around an hour of confusion on receiving the sequencing data, we found the reason behind the odd results - the PyeaR sequence had disappeared from the construct! With no explanation as to how this may have happened, we quickly rethought our plans and returned to the [http://partsregistry.org/Part:BBa_K216009 BBa_K216009] part in order to get some usable characterisation data, and sadly abandoned our attempt to create a PyeaR+LacZ construct.
 "
"