Team:UNIPV-Pavia/Calendar/September/settimana3
From 2010.igem.org
(→September, 16th) |
(→September, 16th) |
||
| (60 intermediate revisions not shown) | |||
| Line 57: | Line 57: | ||
---- | ---- | ||
| - | Inoculum from single colony of | + | Inoculum from single colony of MG42 and MC43 in 5 ml LB+Cm12.5, than ON at 37°C. |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 65: | Line 65: | ||
---- | ---- | ||
| - | I52, I54, I55, I56, I57, I58 plates showed in general few colonies; I52 and | + | I52, I54, I55, I56, I57, I58 plates showed in general few colonies; I52 and I55 showed very few colonies (<=5). Very strange (mumble mumble...), they will be screened ASAP (stored at +4°C). |
Miniprep and quanfification of: | Miniprep and quanfification of: | ||
| Line 76: | Line 76: | ||
*I34: E-X (Vector) | *I34: E-X (Vector) | ||
Gel run/cut of samples (I26 and I31 insert bands were very very soft) | Gel run/cut of samples (I26 and I31 insert bands were very very soft) | ||
| - | [[Image:UNIPV10_14_09_10_gel_run_cut_phasins_2step_final.jpg|thumb| | + | [[Image:UNIPV10_14_09_10_gel_run_cut_phasins_2step_final.jpg|thumb|250px|center| Gel run/cut for digested I26, I31 and I34.]] |
and gel extraction: | and gel extraction: | ||
*I26 (X-P): 2,6 ng/ul | *I26 (X-P): 2,6 ng/ul | ||
| Line 88: | Line 88: | ||
and repeat | and repeat | ||
*I53: I32 (E-S) + I37 (E-X) | *I53: I32 (E-S) + I37 (E-X) | ||
| + | ---- | ||
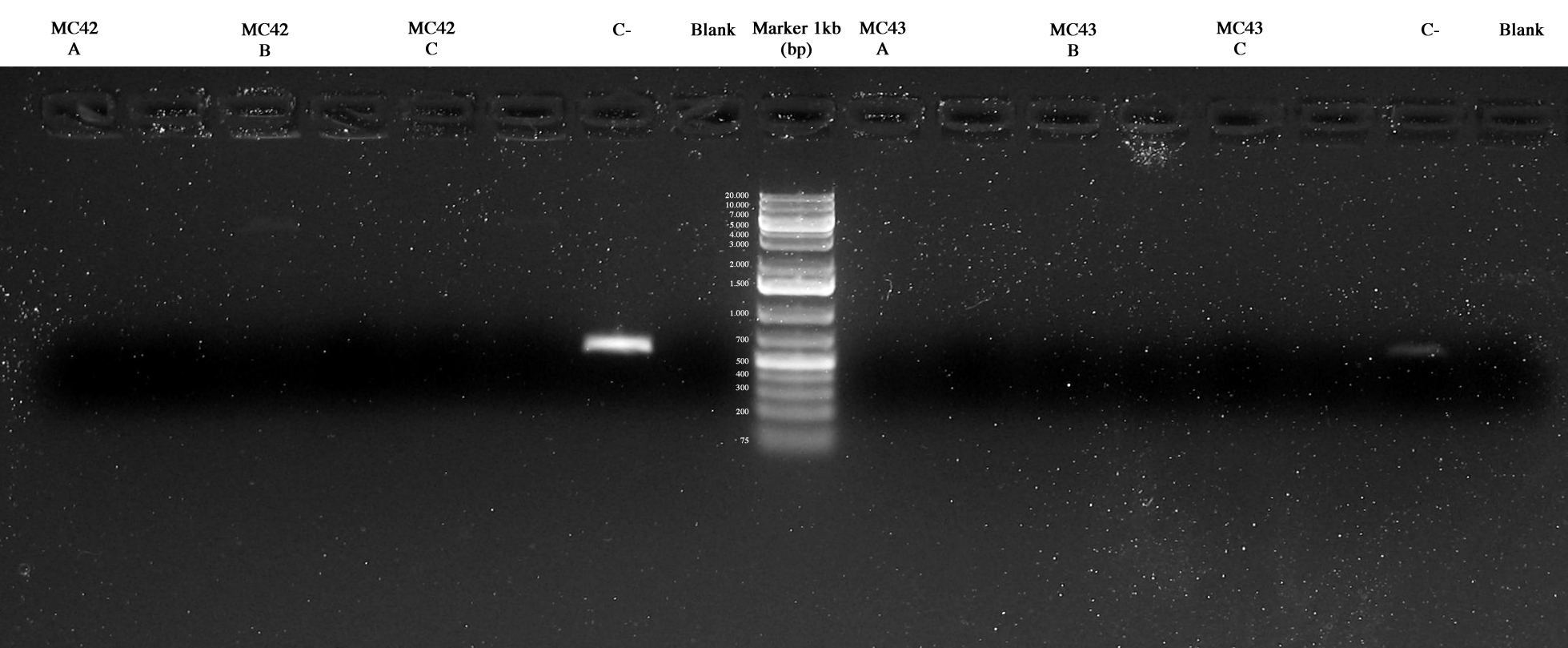
| + | Screening PCR on 3 colonies picked from MC42 and MC43 respectively. | ||
| + | Two method were used: our standard PCR picking a colony from the plate and using it for the PCR and a different protocol picking the colony and treating it with a 95°C 5 min step before the PCR. | ||
| + | |||
| + | The results were the follow: | ||
| + | |||
| + | [[Image:UNIPV1014_09_10_MC42ABC_Cneg_blank_MC43ABC_Cneg_blank.jpg|thumb|300px|center| MC42A/B/C, Cneg, blank, MC43A/B/C, Cneg, blank.]] | ||
| + | |||
| + | Negative controls were ok and maybe MC42_C and MC43_C were ok too but further investigations were necessary. | ||
| - | |||
---- | ---- | ||
| - | + | 5 ul of MG42 and MG43 were streaked on LB+Cm12.5 agar plates and incubated ON at 43°C. | |
| - | + | ||
| + | ---- | ||
| + | |||
| + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test14settembre|Tecan Test]]</font> was performed on prepared samples, after the usual protocol (dilution, medium change and dilution 1:1000). | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==September, 15th== | ==September, 15th== | ||
| - | |||
| - | |||
| - | |||
Transformation of I 53, I59, I60, I61 ligations into ''E. coli'' DH5-alpha. They were plated on LB+Amp agar plates and let grow ON at 37°C. | Transformation of I 53, I59, I60, I61 ligations into ''E. coli'' DH5-alpha. They were plated on LB+Amp agar plates and let grow ON at 37°C. | ||
---- | ---- | ||
| + | 3 colonies were picked from each MG plates and screening PCR was performed also with MC42 and MC43 samples. Two methods with different DNA polymerases were used in order to identify best experimental conditions (Taq polymerase and Pfx accuprime polymerase). The amplicons length (about 4Kb) probably was the reason of our problem in all the PCRs. | ||
| + | |||
| + | [[Image:UNIPV10_15_09_10_PCR_MARKER_MC42ABC_MG42ABC_Cneg_BLANK_MARKER_MG42ABC_MG43ABC_Cneg_BLANK(sopra Taq_sotto Pfx).jpg|thumb|250px|center| Screening PCR. In order: MC42ABC, MG42ABC, Cneg, BLANK, MG42ABC, MG43ABC, Cneg, BLANK (Taq samples over and Pfx samples under)]] | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==September, 16th== | ==September, 16th== | ||
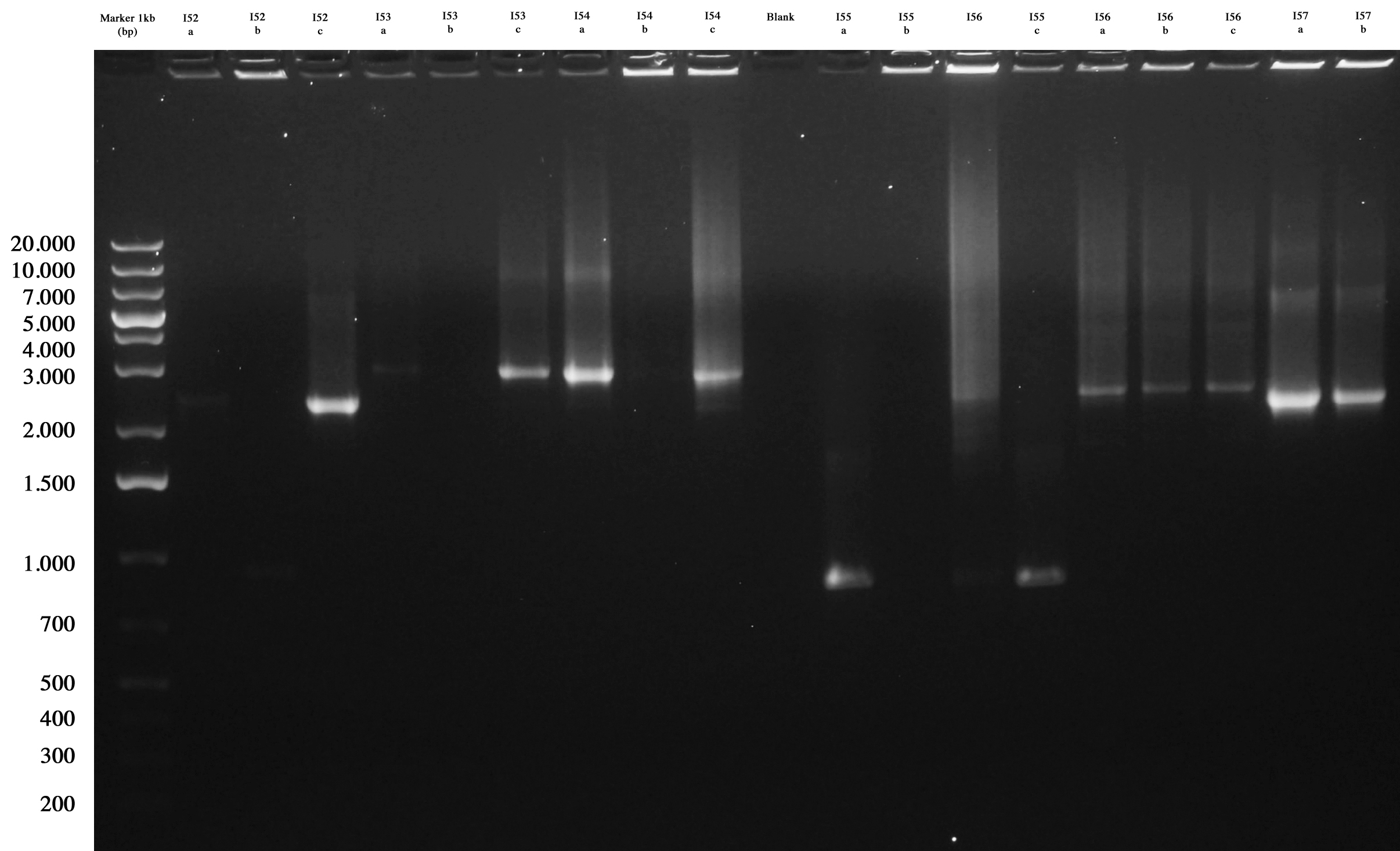
| - | + | All plates showed colonies, so we could perform screening through "colony PCR" for ligations: | |
*I52 | *I52 | ||
*I53 | *I53 | ||
| Line 122: | Line 133: | ||
*I60 | *I60 | ||
*I61 | *I61 | ||
| - | For each plate we picked | + | For each plate we picked 3 colonies that were also inoculated into 1 ml LB+Amp in order to be ready to make glycerol stock of positive ones. |
| - | + | Agarose gel was prepared and samples loaded and run: | |
| - | [[Image:UNIPV10_15_09_10_PCR_colony_ligazioni_fasine_2step.jpg|thumb| | + | {|align="center" |
| - | As you can see ... | + | |[[Image:UNIPV10_15_09_10_PCR_colony_ligazioni_fasine_2step.jpg|thumb|200px|center| Screening gel run for ligations I52..I57.]] || |[[Image:UNIPV10_15_09_10_PCR_colony_ligazioni_fasine_2step2.jpg|thumb|200px|center| Screening gel run for ligations I58..I61.]] |
| + | |} | ||
| + | As you can see I52c, I53a/c, I54a, I56a/b/c, I57a/b are positive. So we decided to make glycerol stocks for I52c, I53c, I54a, I56a and I57a. I55 was negative. We would have performed colony PCR again. Probably all colonies I58..I60 were positive but their length was a little longer than expected. So we decided to further screen them through digestion. | ||
| + | |||
| + | I58a/b/c, I59a/b/c, I60a/b/c and I61a/b/c were let grow ON at 37°C in order to perform miniprep the following day. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | MC and MG cultures OD was referred to the lowest one in order to perform Tecan Test (note that MG42-A didn't grow so we didn't perform test on this sample). Samples were then centrifuged: the resulting pellets were resuspended on 1 ml of PBS. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test16settembre|Tecan Test]]</font> was performed on prepared samples, after the usual protocol (dilution, medium change and dilution 1:1000). | ||
| Line 132: | Line 155: | ||
==September, 17th== | ==September, 17th== | ||
| + | Miniprep and Nanodrop quantification for: | ||
| + | *I58a: 127,5 ng/ul | ||
| + | *I58b: 108,9 ng/ul | ||
| + | *I58c: 100,1 ng/ul | ||
| + | *I59a: 161,6 ng/ul | ||
| + | *I59b: 268,8 ng/ul | ||
| + | *I59c: 214,3 ng/ul | ||
| + | *I60a: 151,6 ng/ul | ||
| + | *I60b: 149,7 ng/ul | ||
| + | *I60c: 157,6 ng/ul | ||
| + | *I61a: 147,1 ng/ul | ||
| + | *I61b: 138,3 ng/ul | ||
| + | *I61c: 125,3 ng/ul | ||
| + | |||
| + | Digestion (for screening) and gel extraction of the positives (for next ligation step) for: | ||
| + | *I58a/b/c: EcoRI-SpeI | ||
| + | *I59a/b/c: XbaI-PstI | ||
| + | *I60a/b/c: E-S | ||
| + | Digestion (only screening) for | ||
| + | *I61a/b/c: E-P | ||
| + | Agarose gel was prepared and samples loaded and run (and cut were necessary) | ||
| + | {|align="center" | ||
| + | |[[Image:UNIPV10_17_09_10_digestioni_screening_I58.jpg|thumb|40px|center| Gel extraction for I58.]] || [[Image:UNIPV10_17_09_10_digestioni_screening_I59-I61.jpg|thumb|200px|center| Screening and gel extraction for positive I59, I60. Screening for I61.]] | ||
| + | |} | ||
| + | As you can see they are all positive (I58 was run a little longer to better separate bands). So we stocked and gel extracted I58c, I59b, I60a and only stocked I61a. | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==September, 18th== | ==September, 18th== | ||
| + | Gel extraction and quantification of: | ||
| + | *I58 (E-S): 13.8 ng/ul | ||
| + | *I59 (X-P): 24.6 ng/ul | ||
| + | *I60 (E-S): 12.8 ng/ul | ||
| + | DNA was than stored at -20°C. | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==September, 19th== | ||
| + | Since there were no more colonies in I55 agar plate, we had to transform it again to perform screening through colony PCR. So I55 was transformed again into ''E. coli'' DH5-alpha and plated on LB+Amp agar plate, that was let grow ON at 37°C. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
<!-- table previous next week --> | <!-- table previous next week --> | ||
Latest revision as of 11:16, 26 October 2010
|
|
||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||
 "
"