Team:Calgary/Project/CpxP
From 2010.igem.org
(New page: {{CalgaryMenu}} <html> <head> <style> div.container{ background-image:url("http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/PageContainer-1.png"); background-repeat: r...) |
|||
| (43 intermediate revisions not shown) | |||
| Line 28: | Line 28: | ||
div.sidebar h1{ | div.sidebar h1{ | ||
color:#7b54cc; | color:#7b54cc; | ||
| + | } | ||
| + | |||
| + | div.sidebar ul{ | ||
| + | font-family:verdana; | ||
} | } | ||
| Line 59: | Line 63: | ||
<div class="container"> | <div class="container"> | ||
| + | |||
<div class="sidebar"> | <div class="sidebar"> | ||
| + | |||
<h1>Project Descriptions</h1> | <h1>Project Descriptions</h1> | ||
| + | |||
<ul> | <ul> | ||
<li><a href="https://2010.igem.org/Team:Calgary/Project/Transcription">Transcription/Translation Reporter Circuit</a></li> | <li><a href="https://2010.igem.org/Team:Calgary/Project/Transcription">Transcription/Translation Reporter Circuit</a></li> | ||
| - | <li>Protein Misfolding Reporters | + | <li><a href="https://2010.igem.org/Team:Calgary/Project/misfolding_overview">Protein Misfolding Reporters</a> |
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Calgary/Project/IbpAB"> | + | <li><a href="https://2010.igem.org/Team:Calgary/Project/IbpAB">Cytoplasmic Stress Detectors</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Calgary/Project/CpxP"> | + | <li><a href="https://2010.igem.org/Team:Calgary/Project/CpxP">Periplasmic Stress Detectors</a></li> |
</ul> | </ul> | ||
</li> | </li> | ||
| + | <li><a href="https://2010.igem.org/Team:Calgary/Project/Controls">Testing Our System</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Calgary/Project/Achievements">Achievements</a></li> | ||
| + | </ul> | ||
</div> | </div> | ||
| Line 77: | Line 87: | ||
<div class="mainbody"> | <div class="mainbody"> | ||
| - | <span id="bodytitle"><h1> | + | <span id="bodytitle"><h1>Periplasmic Stress Detectors</h1></span> |
| + | <br/> | ||
| + | <h2 style="color:#0066CC">What Causes Periplasmic Stress?</h2> | ||
| - | + | <p> Periplasmic stress, also known as envelope stress, is triggered by several factors that influence the ability of the bacteria to communicate with other cells through quorum sensing, pathological pathway, pili formation, outer membrane protein formation which plays a role in adhesion of the cells allowing them to form proper colonies and survive. Many stresses in the periplasmic region are triggered by improper formation of outer protein structures such as pili and lipoprotein disruption that reduces pathogenicity of the bacteria.</p> | |
| - | + | <h2 style="color:#0066CC">Team Calgary circuits for periplasmic stress detector</h2> | |
| + | |||
| + | |||
| + | <table> | ||
| + | <tr><td> | ||
| + | <img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/cpxp-1.png"></img> | ||
| + | <br/> | ||
| + | <img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/cpxr-1.png"></img> | ||
| + | <br/> | ||
| + | <img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/degp-1.png"></img> | ||
| + | </td> | ||
| + | |||
| + | <td> | ||
| + | These images illustrate the promoters of choice by team Calgary for detection of periplasmic stress. The promoters of choice include CpxP, CpxR and DegP promoters. </td></tr></table> | ||
| + | |||
| + | <h2 style="color:#0066CC">How does a native <i>E. coli</i> cell combat periplasmic stress?</h2> | ||
| + | |||
| + | <p> | ||
| + | <i>E coli </i> have several native sigma factors in the genome that are upregulated during envelope stress. Some of the sigma factors include sigma E and sigma 70. These two factors act upon and induce different proteases and chaperones which act to correct the misfolding agent. There is also a new regulon that is currently studied widely, called the Cpx regulon. A well characterized periplasmic regulon found in E. coli is the Cpx regulon. Cpx pathway is involved in maintaining cellular adhesion and keeping periplasmic environment in check. It regulates proteins which are responsible for cellular adhesion and pili formation (Diguiseppe and Silhavy, 2003). Cpx also acts as a periplasmic heat shock pathway. The Cpx pathway is activated by factors which cause protein aggregation in the periplasmic space of gram negative bacteria. Extracellular stresses such as exposure to ethanol, change in pH and temperature change induce the activation of the Cpx heat shock regulon. This increases the transcription of proteins such as CpxP, DegP and CpxR. The Cpx pathway is activated by misfolded proteins or excessive protein concentration in the periplasmic space as well. </p> | ||
| + | |||
| + | <table> <tr><td><a href="http://s872.photobucket.com/albums/ab287/iGEMCalgary_2010/?action=view¤t=Cpxmechanism.png" target="_blank"><img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/Cpxmechanism.png" border="0" alt="Cpx PATHWAY"></a></td> | ||
| + | |||
| + | <td>Figure illustrates Cpx regulon activation pathway. In the lack of stress, the CpxP protein is docked on CpxA transmembrane protein. In the presence of misfolded protein in the envelope, CpxP binds to the misfolded protein which allows CpxA to phosphorylate CpxR which acts as a TF for downstream proteases such as DegP.(Silhavy and Raivio,2001)</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <h3>CpxP</h3> | ||
| + | |||
| + | <p> CpxP is a member of the Cpx regulon which docks on to CpxA, a transmembrane protein normally. But in the case of envelope stress causing protein misfolding in the periplasmic space, CpxP detaches from CpxA and attaches to the misfolded protein in the periplasmic space. </p> | ||
| + | |||
| + | <h3>CpxR</h3> | ||
| + | |||
| + | <p> | ||
| + | CpxR is a transcription factor that is generally dephoshorylated (inactive form). However, in occasion of misfolded protein in the periplasmic space, CpxR is phosphorylated by CpxA which autophosphorylates itself and also has kinase activity towards CpxR. The phosphorylated CpxR then binds to promoter regions that code for proteases such as DegP, ppiD etc. | ||
| + | |||
| + | </p> | ||
| + | <h3>DegP</h3> | ||
| + | |||
| + | <p> DegP is a protease that is activated by CpxR or sigma70 in the event of envelope stress. DegP is transported into the periplasm where it binds to and degrades the misfolded protein. This allows the CpxP protein to be free resulting in binding to CpxA and shutting the Cpx regulon off. | ||
| + | |||
| + | The activation of the Cpx regulon is cyclic in nature and can be reversed when there is a downregulation of activating agents such as misfolded proteins in the cytoplasm. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | <h2 style="color:#0066CC">Circuit usage and sensitivity</h2> | ||
| + | |||
| + | <object width="425" height="344"><param name="movie" value="http://www.youtube.com/v/CO_cK2D4l1s?hl=en&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/CO_cK2D4l1s?hl=en&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="425" height="344"></embed></object> | ||
| + | |||
| + | <p>The circuit that will be constructed to detect protein misfolding will have the CpxP promoter. This has been shown to be the most inducible member of the Cpx regulon. Two other similar circuits will be constructed containing two other Cpx inducible promoter, DegP and CpxR. All three promoters contain binding sites for phosphorylated CpxR, which will induce transcription of our circuit. This allows our promoters to be activated in presence of Cpx inducing signals. Although there is a variety of signals which will activate the cpx cascade, it has been proposed that each of these inducing signals act by creating the accumulation of misfolded proteins. Hence, if periplasmic misfolding occurs in the cell of an E.coli bacteria, the reporter gene, in this case the Red Fluroscent Protein (RFP), will be activated. This provides a simple visual output for protein misfolding within the periplasm of E.coli. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <br/> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td> | ||
| + | |||
| + | <a href="http://s872.photobucket.com/albums/ab287/iGEMCalgary_2010/?action=view¤t=Cpxresponse.png" target="_blank"><img src="http://i872.photobucket.com/albums/ab287/iGEMCalgary_2010/Cpxresponse.png" border="0" alt="Cpx response graph"></a></td> | ||
| + | |||
| + | <td> This diagram illustrated that the CpxP promoter is the most sensitive in the stress regulon. CpxP is followed by CpxR and DegP respectively. These were selected because they are some of the most sensitive in the periplasmic stress detection pathways</td> | ||
| + | |||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <h2 style="color:#0066CC">References</h2> | ||
| + | <p> | ||
| + | DiGiuseppe, P. A., & Silhavy, T. J. (2003). Signal detection and target gene induction by the CpxRA two-component system. Journal of Bacteriology, 185(8), 2432-2440. </p> | ||
| + | <p> | ||
| + | |||
| + | Otto, K., & Silhavy, T. J. (2002). Surface sensing and adhesion of escherichia coli controlled by the cpx-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America, 99(4), 2287-2292. </p> | ||
| + | |||
| + | <p>Raivio T., and Silhavy, T. (2001). Periplasmic stress and ECF Sigma factors. Annual Review of Microbiology, 55, 591–624. | ||
| + | |||
| + | </p> | ||
| - | |||
</div> | </div> | ||
Latest revision as of 03:53, 28 October 2010

Periplasmic Stress Detectors
What Causes Periplasmic Stress?
Periplasmic stress, also known as envelope stress, is triggered by several factors that influence the ability of the bacteria to communicate with other cells through quorum sensing, pathological pathway, pili formation, outer membrane protein formation which plays a role in adhesion of the cells allowing them to form proper colonies and survive. Many stresses in the periplasmic region are triggered by improper formation of outer protein structures such as pili and lipoprotein disruption that reduces pathogenicity of the bacteria.
Team Calgary circuits for periplasmic stress detector

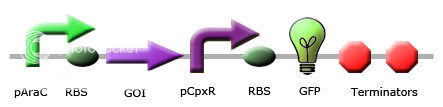

|
These images illustrate the promoters of choice by team Calgary for detection of periplasmic stress. The promoters of choice include CpxP, CpxR and DegP promoters. |
How does a native E. coli cell combat periplasmic stress?
E coli have several native sigma factors in the genome that are upregulated during envelope stress. Some of the sigma factors include sigma E and sigma 70. These two factors act upon and induce different proteases and chaperones which act to correct the misfolding agent. There is also a new regulon that is currently studied widely, called the Cpx regulon. A well characterized periplasmic regulon found in E. coli is the Cpx regulon. Cpx pathway is involved in maintaining cellular adhesion and keeping periplasmic environment in check. It regulates proteins which are responsible for cellular adhesion and pili formation (Diguiseppe and Silhavy, 2003). Cpx also acts as a periplasmic heat shock pathway. The Cpx pathway is activated by factors which cause protein aggregation in the periplasmic space of gram negative bacteria. Extracellular stresses such as exposure to ethanol, change in pH and temperature change induce the activation of the Cpx heat shock regulon. This increases the transcription of proteins such as CpxP, DegP and CpxR. The Cpx pathway is activated by misfolded proteins or excessive protein concentration in the periplasmic space as well.
CpxP
CpxP is a member of the Cpx regulon which docks on to CpxA, a transmembrane protein normally. But in the case of envelope stress causing protein misfolding in the periplasmic space, CpxP detaches from CpxA and attaches to the misfolded protein in the periplasmic space.
CpxR
CpxR is a transcription factor that is generally dephoshorylated (inactive form). However, in occasion of misfolded protein in the periplasmic space, CpxR is phosphorylated by CpxA which autophosphorylates itself and also has kinase activity towards CpxR. The phosphorylated CpxR then binds to promoter regions that code for proteases such as DegP, ppiD etc.
DegP
DegP is a protease that is activated by CpxR or sigma70 in the event of envelope stress. DegP is transported into the periplasm where it binds to and degrades the misfolded protein. This allows the CpxP protein to be free resulting in binding to CpxA and shutting the Cpx regulon off. The activation of the Cpx regulon is cyclic in nature and can be reversed when there is a downregulation of activating agents such as misfolded proteins in the cytoplasm.
Circuit usage and sensitivity
The circuit that will be constructed to detect protein misfolding will have the CpxP promoter. This has been shown to be the most inducible member of the Cpx regulon. Two other similar circuits will be constructed containing two other Cpx inducible promoter, DegP and CpxR. All three promoters contain binding sites for phosphorylated CpxR, which will induce transcription of our circuit. This allows our promoters to be activated in presence of Cpx inducing signals. Although there is a variety of signals which will activate the cpx cascade, it has been proposed that each of these inducing signals act by creating the accumulation of misfolded proteins. Hence, if periplasmic misfolding occurs in the cell of an E.coli bacteria, the reporter gene, in this case the Red Fluroscent Protein (RFP), will be activated. This provides a simple visual output for protein misfolding within the periplasm of E.coli.
References
DiGiuseppe, P. A., & Silhavy, T. J. (2003). Signal detection and target gene induction by the CpxRA two-component system. Journal of Bacteriology, 185(8), 2432-2440.
Otto, K., & Silhavy, T. J. (2002). Surface sensing and adhesion of escherichia coli controlled by the cpx-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America, 99(4), 2287-2292.
Raivio T., and Silhavy, T. (2001). Periplasmic stress and ECF Sigma factors. Annual Review of Microbiology, 55, 591–624.
 "
"

