Team:UNAM-Genomics Mexico/Modeling
From 2010.igem.org
| (4 intermediate revisions not shown) | |||
| Line 69: | Line 69: | ||
image:UNAM-Genomics_Mexico_Green_emission_phots.jpg|Data for Green Emission system with photons per second | image:UNAM-Genomics_Mexico_Green_emission_phots.jpg|Data for Green Emission system with photons per second | ||
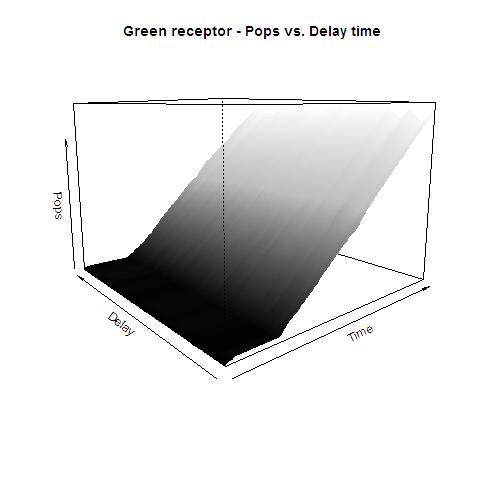
image:UNAM-Genomics_Mexico_Green_receptor_Pop.jpg|Data for Green Receptor system with absolute polymerases | image:UNAM-Genomics_Mexico_Green_receptor_Pop.jpg|Data for Green Receptor system with absolute polymerases | ||
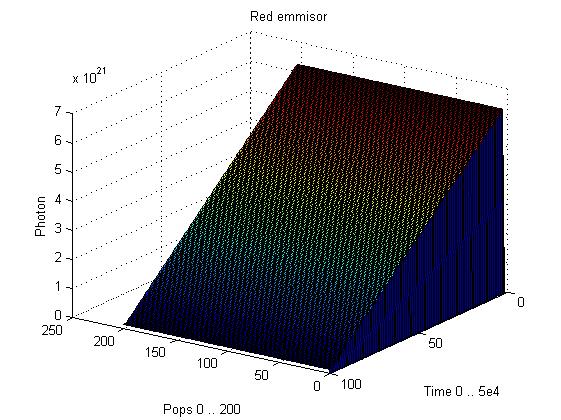
| + | image:UNAM-Genomics_Mexico_Red_emmision.jpg|Data for the Red emission system with total photons | ||
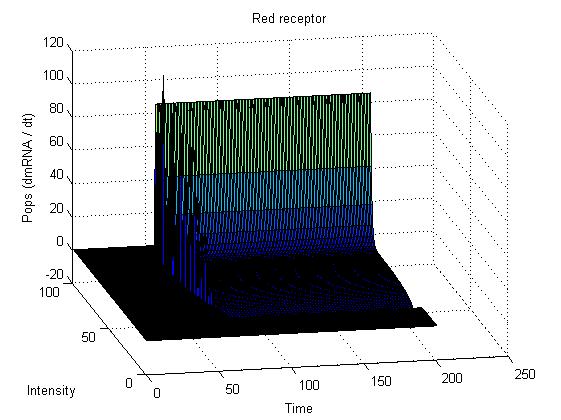
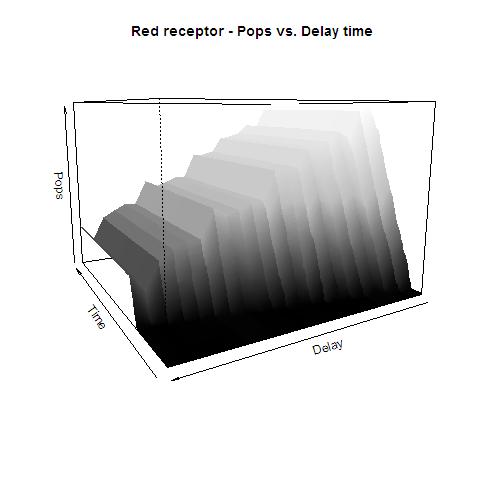
| + | image:UNAM-Genomics_Mexico_Red_receptor.jpg|Data for the Red receptor system with Polymersases per second | ||
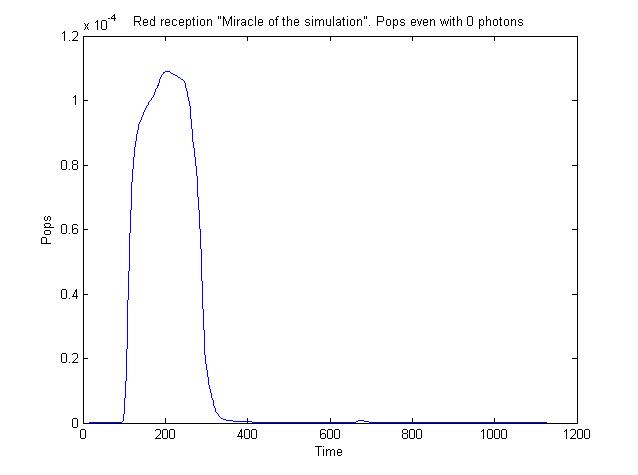
image:UNAM-Genomics_Mexico_Miracle.jpg|Aberrant behavior with red receptor... | image:UNAM-Genomics_Mexico_Miracle.jpg|Aberrant behavior with red receptor... | ||
</gallery> | </gallery> | ||
| + | |||
| + | Extra code to derivate the absolute data into flux, to organize and other miscelaneous functions can be found [[media:UNAM-Genomics_Mexico_FINAL.zip|here]]. | ||
We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner. | We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner. | ||
| Line 99: | Line 103: | ||
The kapppa code is [[media:UNAM-Genomics_Mexico_Green_Receptor_Kappa.ka.txt|here]], and the perl runner is [[media:UNAM-Genomics_Mexico_Green_Batch.txt|this]] one. | The kapppa code is [[media:UNAM-Genomics_Mexico_Green_Receptor_Kappa.ka.txt|here]], and the perl runner is [[media:UNAM-Genomics_Mexico_Green_Batch.txt|this]] one. | ||
| + | |||
====Red Receptor==== | ====Red Receptor==== | ||
| Line 105: | Line 110: | ||
The kapppa code is [[media:UNAM-Genomics_Mexico_Red_Receptor_Kappa.ka.txt|here]], and the perl runner is [[media:UNAM-Genomics_Mexico_Red_Batch.txt|this]] one. | The kapppa code is [[media:UNAM-Genomics_Mexico_Red_Receptor_Kappa.ka.txt|here]], and the perl runner is [[media:UNAM-Genomics_Mexico_Red_Batch.txt|this]] one. | ||
| + | |||
| + | |||
| + | ====Emission==== | ||
| + | |||
| + | Our emission models reveal the following behavior, even with abysmal levels of PopS, the Photons/second output is massive. Unfortunately, we didn't have time to adjust the parameters for these graphics to generate the 3D plots. | ||
| + | |||
===Neural Network=== | ===Neural Network=== | ||
| Line 124: | Line 135: | ||
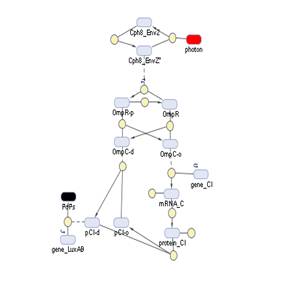
Had we been able to get Cph8 working, we would have found that the OmpR-P concentration was saturating the available plasmid slots, even with the kinase knocked off it was always superior to 120 molecules per cell. This behavior was revealed by our model, so for the construction we would recommend a high-copy number plasmid. | Had we been able to get Cph8 working, we would have found that the OmpR-P concentration was saturating the available plasmid slots, even with the kinase knocked off it was always superior to 120 molecules per cell. This behavior was revealed by our model, so for the construction we would recommend a high-copy number plasmid. | ||
| + | |||
| + | |||
| + | ===Other Modeling Ideas=== | ||
| + | |||
| + | We also though of modeling a bio-cable. The object of this was to determine if our bacteria had the characteristics of a quaternary single-channel communication platform, or if it behaved instead as a 4 binary channels array. its use would be to use light to communicate one edge of the bio-cable by a single input, therefore have automatic transfer of light to the other extreme of it. | ||
| + | The Collaboration with the other UNAM team was along these tracks: modeling this behavior. However, due to a general lack of time, results are inconclusive... | ||
}} | }} | ||
Latest revision as of 03:57, 28 October 2010

Modeling 101
We wanted to simulate our system in silico to answer some questions and to explore the dynamics of our systems. Moreover, since we wanted to characterize and refine the transfer function for our interfaces, from photon flux to Pop flux, we chose to simulate our system as 6 individual modules.
We chose to use [http://www.mathworks.com/products/simbiology/ Simbiology] as a modeling platform in order to take advantage of the implemented solvers in it. Eventually we realized that we wanted a stochastic simulation. So we took a look at the [http://www.kappalanguage.org/ Kappa] platform. In our search for ways to use the Kappa syntax, and having trouble with the RuleBase.org edition, we joined the [http://groups.google.com/group/kappa-users KappaUsers] forum and made a [http://groups.google.com/group/kappa-users/browse_thread/thread/c77a720b49e4934 Tutorial] on .ka scripts.
The Model, Original Idea
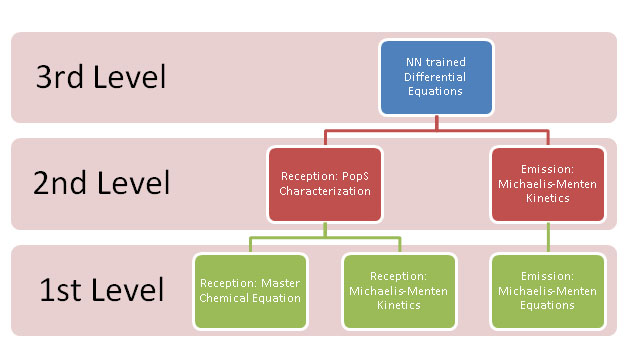
We decided to try some different approaches at modeling. At first, we were planning on a three level implementation where we could fall back on simpler levels should experimental data be scarce. The levels are thus:
The most complex level was the third, where we would have ODEs trained by a Neural Network (multilayer, minimal error, perceptron) describing the system's behavior. However, to do this, we needed the working system. In absence of this, we fell back to the second level.
The second level required the PopS characterization, however it used existing models (notably mass action kinetics) to simulate the rest of the system. In absence of PopS characterization, we fell to the first level.
The basic level required nothing but what should be found-able in the literature. We did found enough parameters to do the Simbiology model, however we didn't found the required for Kappa. In theory, we could have generated said stochastic parameters from the deterministic ones, but these required an extreme amount of computational time.
The Real Model
So, nearing the WikiFreeze craze, we decided to simply stuff. We got rid of the differential equations, and instead focused on using the existing implementations of the Master Chemical Equation found within Simbiology and Kappa to simulate our model. For simpler dynamics, we simulated with mass-actions kinetics.
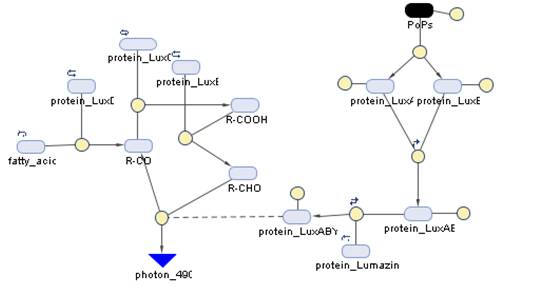
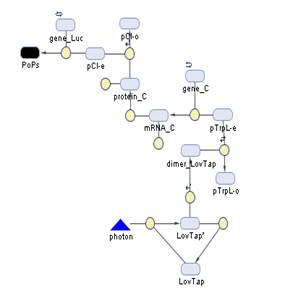
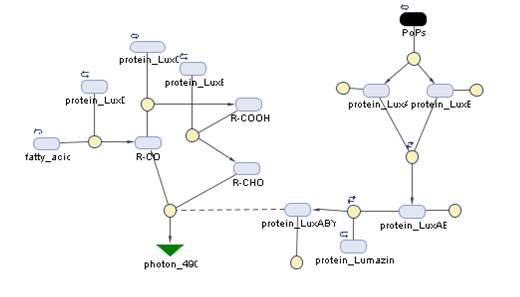
The Systems
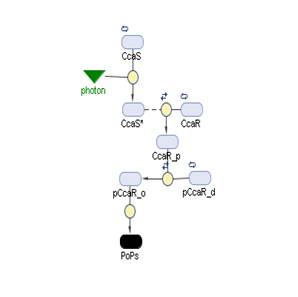
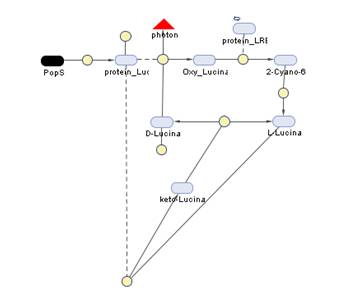
The Simbiology maps:
Simbiology
So having Simbiolgy, we decided to model our interfaces using it's deterministic solvers. Some results are here. We would include the original .sbproj files if they the Server didn't impose a file size cap.
Extra code to derivate the absolute data into flux, to organize and other miscelaneous functions can be found here.
We chose to simulate the flux of output given a determined input. So we have for a wide range of inputs, the resultant outputs in a time manner.
Kappa
Also, having the Kappa binaries, we decided to model the system using Kappa as a stochastic modeling platform. Some preliminary results would be here if the Server didn't have an unexplainable aversion for .7z, .rar, or partial intolerance for .zip As soon as the Server decides to tolerate our files, we will upload them. In the mean time, here are some sample data. Please take note of the file extensions, as due to server issues, we had to append a .txt extension to upload the original code.
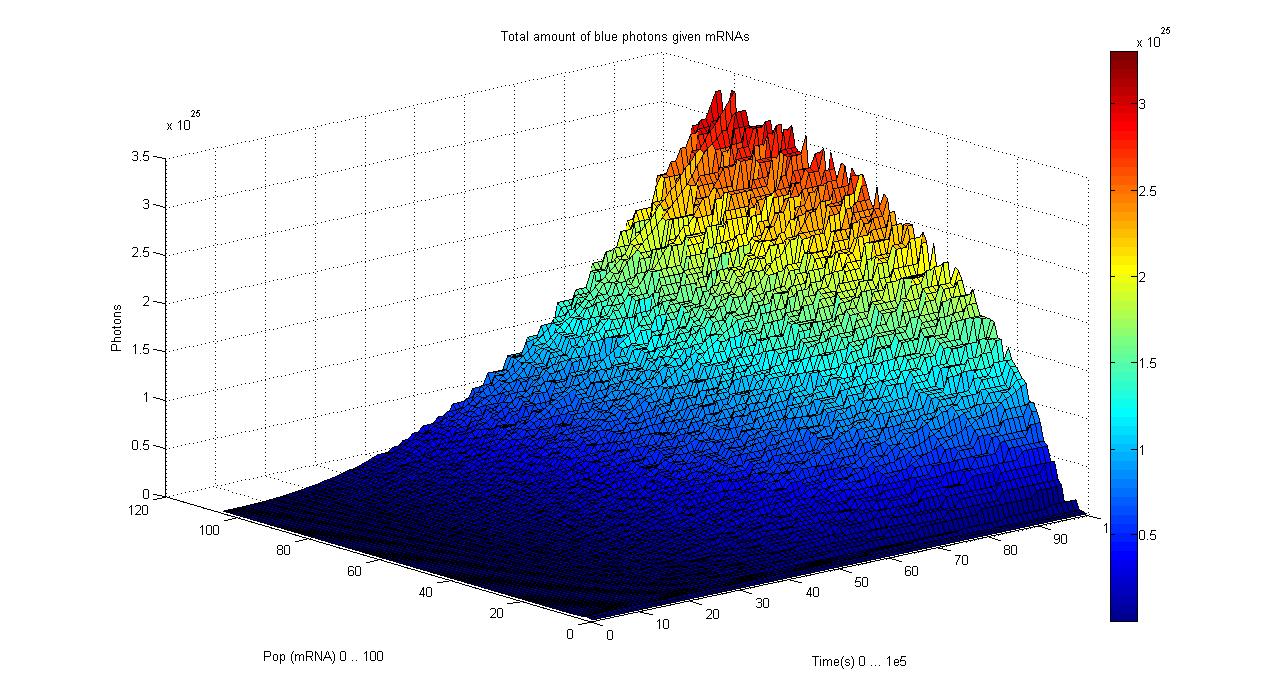
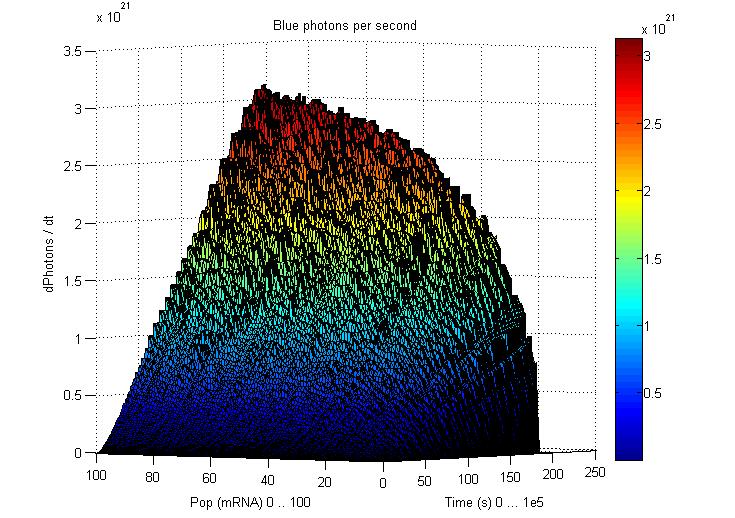
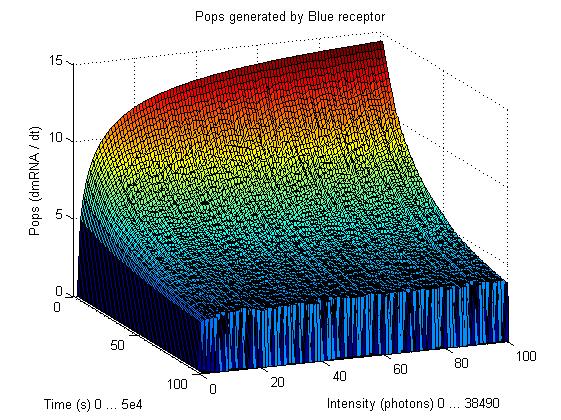
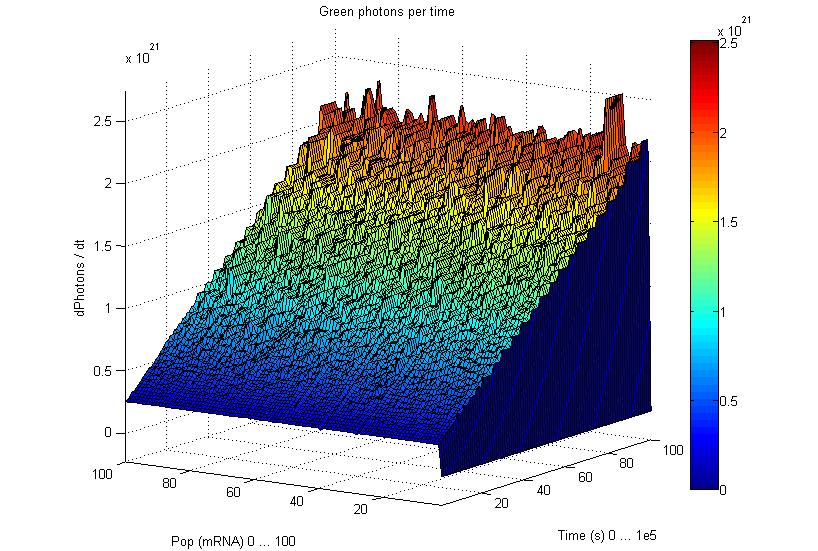
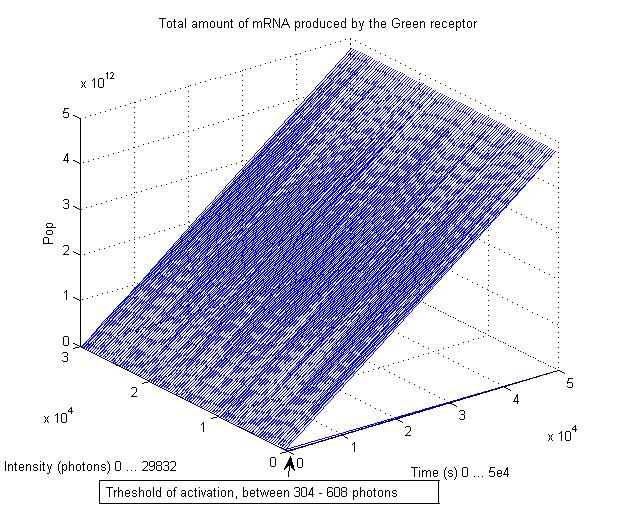
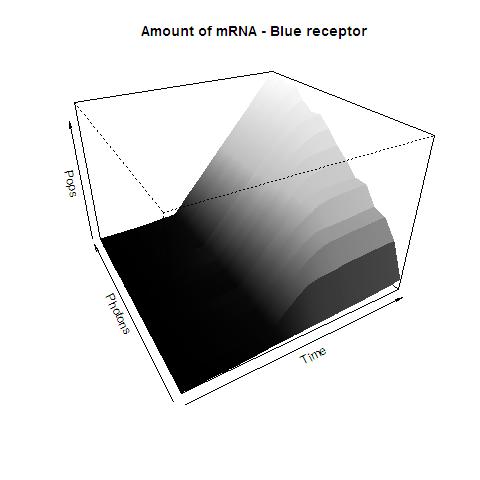
This graphs plot the Pop output of our Receptors, notice this output is of a TOTAL final production, not the rate of production (i.e. Polymerases per Second vs. Total Amount of Polymerases). The .ka code is available, however take into account that said code generates a single plane, to generate the 3D plot we used a perl script. The system begins in dark state, and at a given time (you can guess which one), there's a photon influx. These graphs are however in arbitrary units. We wanted to make a more useful characterization, but these simulations required some extreme computing time.
About the files, since the Server has .ka indigestion, we changed the file extensions to .txt, idem for the perl scripts. These generate a data_final file, which we then imported into R and rendered into a perspective plot.
The blue and red receptors show the dramatic Pop production correlating with the photon input. The green receptor shows a toggle-switch activity, this is due to the photo-conversion phenomenon. The system will cease to produce PopS when struck by a contrary photon (far red in this case).
Blue Receptor
The kapppa code is here, and the perl runner is this one.
Green Receptor
The kapppa code is here, and the perl runner is this one.
Red Receptor
The kapppa code is here, and the perl runner is this one.
Emission
Our emission models reveal the following behavior, even with abysmal levels of PopS, the Photons/second output is massive. Unfortunately, we didn't have time to adjust the parameters for these graphics to generate the 3D plots.
Neural Network
In Training... Having over a million data to map, our Levenberg-Marquandt algorithm is taking longer than expected.
Conclusions
A model that says nothing is not very useful. However, our model has revealed some very interesting phenomena.
Photomultiplier
All our simulations, data, and intuition show that our system will behave as a photomultiplier. We are very excited at this, since this is the very first synthetic-organic photomultiplier device.
Construction Issue
Had we been able to get Cph8 working, we would have found that the OmpR-P concentration was saturating the available plasmid slots, even with the kinase knocked off it was always superior to 120 molecules per cell. This behavior was revealed by our model, so for the construction we would recommend a high-copy number plasmid.
Other Modeling Ideas
We also though of modeling a bio-cable. The object of this was to determine if our bacteria had the characteristics of a quaternary single-channel communication platform, or if it behaved instead as a 4 binary channels array. its use would be to use light to communicate one edge of the bio-cable by a single input, therefore have automatic transfer of light to the other extreme of it.
The Collaboration with the other UNAM team was along these tracks: modeling this behavior. However, due to a general lack of time, results are inconclusive...iGEM
iGEM is the International Genetically Engineered Machines Competition, held each year at MIT and organized with support of the Parts Registry. See more here.Synthetic Biology
This is defined as attempting to manipulate living objects as if they were man-made machines, specifically in terms of genetic engineering. See more here.Genomics
We are students on the Genomic Sciences program at the Center for Genomic Sciences of the National Autonomous University of Mexico, campus Morelos. See more here.This site is best viewed with a Webkit based Browser (eg: Google's Chrome, Apple's Safari),
Trident based (Microsoft's Internet Explorer) or Presto based (Opera) are not currently supported. Sorry.
 "
"