Team:SDU-Denmark/K343007
From 2010.igem.org
(Difference between revisions)
(→Growth assay) |
|||
| (4 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
=== Growth of bacterial culture on semi-solid agar plates=== | === Growth of bacterial culture on semi-solid agar plates=== | ||
==== Experiment 1 ==== | ==== Experiment 1 ==== | ||
| - | The bacteria and plates were prepared after our own protocol, which you can find here: [https://2010.igem.org/Team:SDU-Denmark/protocols#Photosensor_characterisation PS1.1] <br> [[Image:Ingeniør1.JPG|200px|Wildtype bacteria (Left half exposed to light, right half exposed to darkness)|thumb|'''Figure 1''': ''E. coli'' MG1655 exposed to blue light on the left half and to darkness on the right half.]][[Image:Ingeniør4.JPG|200px|Phototactic bacteria (Left half exposed to light, right half exposed to darkness)|thumb|'''Figure 2''': ''E. coli'' MG1655-pSB3T5-K343007 exposed to blue light on the left half and to darkness on the right half.]] | + | The bacteria and plates were prepared after our own protocol, which you can find here: protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#Photosensor_characterisation PS1.1] <br> [[Image:Ingeniør1.JPG|200px|Wildtype bacteria (Left half exposed to light, right half exposed to darkness)|thumb|'''Figure 1''': ''E. coli'' MG1655 exposed to blue light on the left half and to darkness on the right half.]][[Image:Ingeniør4.JPG|200px|Phototactic bacteria (Left half exposed to light, right half exposed to darkness)|thumb|'''Figure 2''': ''E. coli'' MG1655-pSB3T5-K343007 exposed to blue light on the left half and to darkness on the right half.]] |
| - | Since exposure to blue light should decrease the phototaxic bacterias tumbling frequency, the expected result was that the colony which was placed between the light and dark half of the plate would spread out in the darkness and would not move further when it reached the light. This is counter-intuitive, since decreased tumbling should lead to a longer distance traveled. What happens at the microscopic scale in semisolid agar is that the agar creates a matrix like structure where | + | Since exposure to blue light should decrease the phototaxic bacterias tumbling frequency, the expected result was that the colony which was placed between the light and dark half of the plate would spread out in the darkness and would not move further when it reached the light. This is counter-intuitive, since decreased tumbling should lead to a longer distance traveled. What happens at the microscopic scale in semisolid agar is that the agar creates a matrix like structure where channels are running through the agar, that the bacteria can swim through. The decrease in tumbling frequency of the bacteria will make it harder for them to find the channels in the agar to swim through, which leads to them being trapped where they were placed. The result is that a colony which shows an increased run time, will look as if it was non-motile on these plates [[https://2010.igem.org/Team:SDU-Denmark/project-p#References 1]]. Our results showed exactly this; the bacterial culture had spread out onto the dark half of the plate and did not get nearly as far on the half exposed to light. This experiment was done with a normal wildtype MG1655 and a non-motile strain of ''E.coli'', DH5alpha, as controls. As expected these cells did not show anything like the behavior described above, which indicates that the effect stems from the modification of our photosensing bacteria. |
<br> | <br> | ||
<br> | <br> | ||
| Line 17: | Line 17: | ||
==== Experiment 2==== | ==== Experiment 2==== | ||
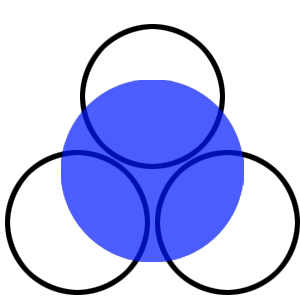
| - | The experiment described above was repeated in a more controlled environment. This means that there were no changes as to how the plates or bacterial cultures were prepared, so we refer again to [https://2010.igem.org/Team:SDU-Denmark/protocols#Photosensor_characterisation PS1.1] in regards to how this was done. The difference lies in the setup of the light-controlled environment. What we did this time was that the plates was illuminated from above by a single light source in an otherwise completely dark environment, we prepared a cut-out so that the light would only hit one half of our plates and the other half would remain in the dark. Since the problem with the last experiment was that the light source was just normal white light (which contains a lot of different wavelengths), this time around we used an optical filter so that only light with a wavelength of around 470nm could pass through, which resulted in a blue light shining down on the plates. The light-source itself was a run-of-the-mill flashlight, with a blue light filter installed in front of the lens. To eliminate the effect of temperature gradients inside the incubator, the three samples were placed in a triangle formation in the center of the | + | The experiment described above was repeated in a more controlled environment. This means that there were no changes as to how the plates or bacterial cultures were prepared, so we refer again to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#Photosensor_characterisation PS1.1] in regards to how this was done. The difference lies in the setup of the light-controlled environment. What we did this time was that the plates was illuminated from above by a single light source in an otherwise completely dark environment, we prepared a cut-out so that the light would only hit one half of our plates and the other half would remain in the dark. Since the problem with the last experiment was that the light source was just normal white light (which contains a lot of different wavelengths), this time around we used an optical filter so that only light with a wavelength of around 470nm could pass through, which resulted in a blue light shining down on the plates. The light-source itself was a run-of-the-mill flashlight, with a blue light filter installed in front of the lens. To eliminate the effect of temperature gradients inside the incubator, the three samples were placed in a triangle formation in the center of the designed light box. <br> |
| - | [[Image:Lightbox5k.gif|150px|thumb|left| Schematic over plate positioning and areas exposed to light.]] | + | [[Image:Lightbox5k.gif|150px|thumb|left|'''Figure 3:''' Schematic over plate positioning and areas exposed to light.]] |
<br> | <br> | ||
| - | Another deviation from the protocol is that the | + | Another deviation from the protocol is that the cultures were not spotted onto the center of the plate, but instead one drop of culture was placed in the center of the light exposed half and another drop (of the same culture) was spotted onto the dark half. This would give us more information on how the bacteria would behave when directly exposed to either the darkness or illuminated surroundings, instead of the gradient that was present in the first experiment.<br> |
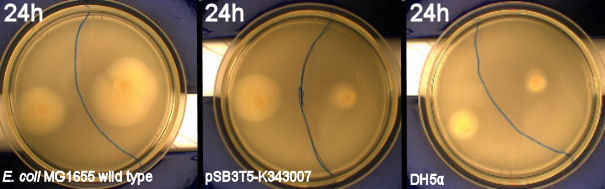
| - | From the results of the first experiment we expected the culture containing K343007 to spread out in the dark and not to spread when exposed to light. The | + | From the results of the first experiment we expected the culture containing K343007 to spread out in the dark and not to spread when exposed to light. The wild type bacteria should spread out evenly no matter if it is exposed to light or not and the non-motile strain (DH5alpha) should not move regardless of the light conditions. Our expectations from the first experiment were fulfilled, as the bacteria behaved exactly as expected. This made it possible to conclude that the photosensor has an effect on the bacteria's tumbling frequency. Whether it actually reduces the tumbling is not possible to say, since both an increased and reduced tumbling frequency will look alike.<br><br> |
''From left to right: MG1655 (Wildtype), Photosensor (Bacteria containing K343007), DH5alpha (non-motile strain) | ''From left to right: MG1655 (Wildtype), Photosensor (Bacteria containing K343007), DH5alpha (non-motile strain) | ||
| - | [[Image:Team-sdu-denmark-PS-lightbox-geneous.jpg|630px|center|thumb]]<br> | + | [[Image:Team-sdu-denmark-PS-lightbox-geneous.jpg|630px|center|thumb|'''Figure 4:''' wild type E. coli strain MG1655 and MG1655-pSB1C3-K343007 grown on motility agar (LB media + 0.3% agarose) incubated at 37 degrees celcius for 24 hours. Plates were placed with one part exposed to light and the other part in darkness. A difference in growth of the transformed bacteria exposed to light contra the transformants that grew in darkness is clearly seen. The cells grown in light did not spread as much as the ones grown in darkness. The wild type seems to grow best when exposed to light while DH5alpha grows a little better in darkness]]<br> |
After this we could conclude that the part had an impact on the bacterial motility, but we had to find out which. This lead us to our next experiment, which was intended for determining what happened to the tumbling frequency:<br> | After this we could conclude that the part had an impact on the bacterial motility, but we had to find out which. This lead us to our next experiment, which was intended for determining what happened to the tumbling frequency:<br> | ||
<br> | <br> | ||
=== Videomicroscopy and computer analysis of bacterial motility === | === Videomicroscopy and computer analysis of bacterial motility === | ||
| - | To get an exact idea of what is happening to the bacteria | + | To get an exact idea of what is happening to the bacteria expressing the photosensor we decided on doing video microscopy of motile bacteria. The bacterial cultures were prepared according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#PS1.1 PS1.1]. Wildtype MG1655 and DH5alpha was used as controls. |
The actual microscopy was done on a Nikon eclipse TE2000-S microscope with an optical magnification of 1000x. <br> | The actual microscopy was done on a Nikon eclipse TE2000-S microscope with an optical magnification of 1000x. <br> | ||
| - | To be able to observe an effect in the modified bacteria we had to expose the cultures to light, which was done through the microscope's integrated light source, which could switch between blue, green and ambient light. To ensure that the blue light was at the correct wavelength the same optical filter that was used for the plate experiments was inserted between the lightsource and the microscopy slide. Since microscopy is impossible without light, we had to use red light as a replacement for darkness, the source of the red light was ambient light from the microscope's light source filtered through a red optical filter (about 580nm). This should not have any effect on the phototactic bacteria according to [[https://2010.igem.org/Team:SDU-Denmark/project-p#References 2]]. The room where the experiment was carried out was kept as dark as possible, as to eliminate the chance of the phototactic bacteria being | + | To be able to observe an effect in the modified bacteria we had to expose the cultures to light, which was done through the microscope's integrated light source, which could switch between blue, green and ambient light. To ensure that the blue light was at the correct wavelength the same optical filter that was used for the plate experiments was inserted between the lightsource and the microscopy slide. Since microscopy is impossible without light, we had to use red light as a replacement for darkness, the source of the red light was ambient light from the microscope's light source filtered through a red optical filter (about 580nm). This should not have any effect on the phototactic bacteria according to [[https://2010.igem.org/Team:SDU-Denmark/project-p#References 2]]. The room where the experiment was carried out was kept as dark as possible, as to eliminate the chance of the phototactic bacteria being exposed to light. The actual experiment was carried out in the following manner:<br><br> |
| - | + | # Expose bacteria to red light for 1 minute, while recording video.<br> | |
| - | + | # Move to another spot on the slide, repeat 1. (repeated until 5 videos were recorded)<br> | |
| - | + | # Expose bacteria to blue light for 1 minute, while recording video.<br> | |
| - | + | # Wait for 30 seconds, for the bacteria to reset themselves.<br> | |
| - | + | # Move the focus to another spot on the slide, repeat 3. (x5)<br><br> | |
This was done with all three strains of bacteria.<br> | This was done with all three strains of bacteria.<br> | ||
An example of the results: <br><html> | An example of the results: <br><html> | ||
<object width="320"><param name="movie" value="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="300"></embed></object> <object width="320"><param name="movie" value="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="320" ></embed></object> </html><br><br> | <object width="320"><param name="movie" value="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="300"></embed></object> <object width="320"><param name="movie" value="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="320" ></embed></object> </html><br><br> | ||
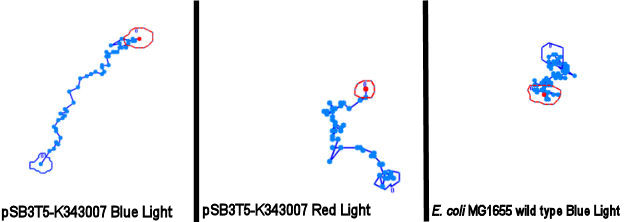
| - | When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor | + | When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor containing bacteria seem to show slightly increased motility when exposed to blue light. Even though the bacteria displayed disappointing motility, we went on and did computer analysis on the videos by the help of the open source software [http://db.cse.ohio-state.edu/CellTrack/ "CellTrack"]. The paths that were mapped showed a longer traveling distance for the bacteria expressing the photosensor compared to the rest of the samples when exposed to blue light. We picked 10 bacteria from each sample, which amounts to 60 bacteria that were tracked. This is a rather small amount, but because of the time-intensive procedure a more thorough data analysis was not possible.<br><br> |
''Samples of the tracked paths (from left to right): Photosensor exposed to blue light, photosensor exposed to red light, wildtype exposed to blue light.'' <br> | ''Samples of the tracked paths (from left to right): Photosensor exposed to blue light, photosensor exposed to red light, wildtype exposed to blue light.'' <br> | ||
| - | [[Image:Team SDU-Denmark-PS-traels-figur.jpg|630px|center|thumb]]<br><br> | + | [[Image:Team SDU-Denmark-PS-traels-figur.jpg|630px|center|thumb|'''Figure 5:''' Tracking of ''E. coli'' strain MG1655-pSB3T5 exposed to blue light, MG1655-pSB3T5 exposed to red light and wild type MG1655 exposed to blue ligt. Both transformants move straighter than the wild type. However, the streightest path is seen when cells are exposed to blue light]]<br><br> |
| - | This might indicate a decreased tumbling frequency, but the low motility of the bacteria, the inadequacy of the cell track software and the small amount of data | + | This might indicate a decreased tumbling frequency, but the low motility of the bacteria, the inadequacy of the cell track software and the small amount of data analyzed make results of this experiment unreliable and a new protocol had to be devised.<br><br> |
===Computerized analysis of the bacterial motility with the THOR prototype by [http://unisensor.dk/ Unisensor A/S] and the Unify software=== | ===Computerized analysis of the bacterial motility with the THOR prototype by [http://unisensor.dk/ Unisensor A/S] and the Unify software=== | ||
| - | For this experiment we changed our protocol for cultivating swimming bacteria in order to optimize their | + | For this experiment we changed our protocol for cultivating swimming bacteria in order to optimize their motility [https://2010.igem.org/Team:SDU-Denmark/protocols#PS1.2 PS1.2]<br> |
| - | The machine and software used for the analysis are prototypes and still under heavy development. Because of trade secrets it is impossible for us to explain how the machine works or give a detailed explanation of it's mechanism until the THOR has reached production status. Simplistically said it is a very advanced video microscope, with which it is possible to | + | The machine and software used for the analysis are prototypes and still under heavy development. Because of trade secrets it is impossible for us to explain how the machine works or give a detailed explanation of it's mechanism until the THOR has reached production status. Simplistically said it is a very advanced video microscope, with which it is possible to analyze liquids and the particles inside those in both 2D and 3D, while also tracking them over time and also space, since the camera/optical lens can also move along the slide, while recording. <br> |
| - | 2.5 | + | 2.5 µl of the bacterial dilution were placed on the center of the microscopy slide and covered by a cover slip, which results in a layer of liquid with a height of 6 µm. Afterwards the sample was sealed with mineral oil as to prevent a flow in the liquid. The data collection was done exactly as described in the previous experiment, with the exception that we used diodes instead of optical filters. The light used for recording was a red LED diode with a wavelength of 660 nm. The light source used for the exposure was a blue light LED, which emitted light at a wavelength of 470 nm, and hit the sample with an angle of attack of 17°. Moreover we also recorded the bacteria's behavior when exposed to a blue light gradient and varying intensities of light measured in milliampere ranging from 1mA to 20mA. All the measurements were carried out on the modified phototactic bacteria and the wildtype MG1655 worked as a control.<br> |
The resulting videos looked like these examples:<br> | The resulting videos looked like these examples:<br> | ||
<br><html> | <br><html> | ||
| Line 57: | Line 57: | ||
<br></html> | <br></html> | ||
The bacteria that are not moving in the video are assumed to be sticking to the glass or coverslip.<br> | The bacteria that are not moving in the video are assumed to be sticking to the glass or coverslip.<br> | ||
| - | From these videos we extracted data through Unisensor's in-house programmed software Unify and it's tracking extension. For each frame the bacteria were identified and their location compared to that of the frame before and thereby, tracking the bacteria over time. The resulting data which served as output for each frame was: Location of bacteria given as coordinates (X,Y), bacterial orientation in angle ranging from -180° to 180° and length of the path was tracked in pixels. An example of the unify | + | From these videos we extracted data through Unisensor's in-house programmed software Unify and it's tracking extension. For each frame the bacteria were identified and their location compared to that of the frame before and thereby, tracking the bacteria over time. The resulting data which served as output for each frame was: Location of bacteria given as coordinates (X,Y), bacterial orientation in angle ranging from -180° to 180° and length of the path was tracked in pixels. An example of the unify software analysing the paths of the bacteria and at the same creating a video of the individual tracks is showed below:<br> |
<html><object width="425" height="344"><param name="movie" value="http://www.youtube.com/v/lARwMFCay_Q?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/lARwMFCay_Q?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="425" height="344" align="center"></embed></object></html><br> | <html><object width="425" height="344"><param name="movie" value="http://www.youtube.com/v/lARwMFCay_Q?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/lARwMFCay_Q?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="425" height="344" align="center"></embed></object></html><br> | ||
| - | Here is an example of a .txt file containing the results: [https://static.igem.org/mediawiki/2010/6/6b/TrackResult_iGEM-WTred1.txt Tracks of wildtype bacteria exposed to red light]. If interested, please request the whole dataset from us, which will be sent out as a | + | Here is an example of a .txt file containing the results: [https://static.igem.org/mediawiki/2010/6/6b/TrackResult_iGEM-WTred1.txt Tracks of wildtype bacteria exposed to red light]. If interested, please request the whole dataset from us, which will be sent out as a 450MB zip file.<br> |
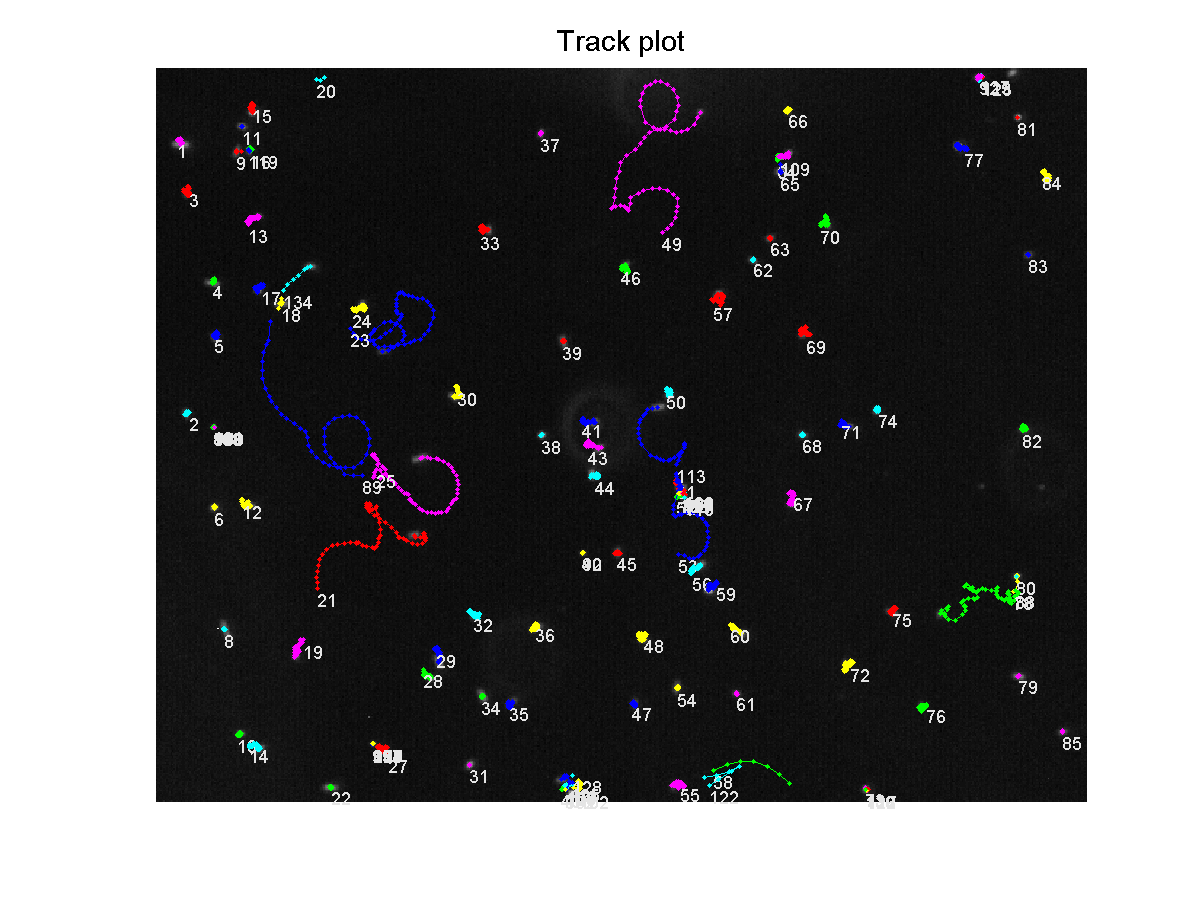
From this file, a trackplot was generated, showing each bacteria's path over the recorded time:<br> | From this file, a trackplot was generated, showing each bacteria's path over the recorded time:<br> | ||
| - | [[Image:TrackPlotTagged.png|thumb|400px|center|Bacteria containing K343007 tracked over 7.5 seconds]]<br> | + | [[Image:TrackPlotTagged.png|thumb|400px|center|'''Figure 6:''' Bacteria containing K343007 tracked over 7.5 seconds]]<br> |
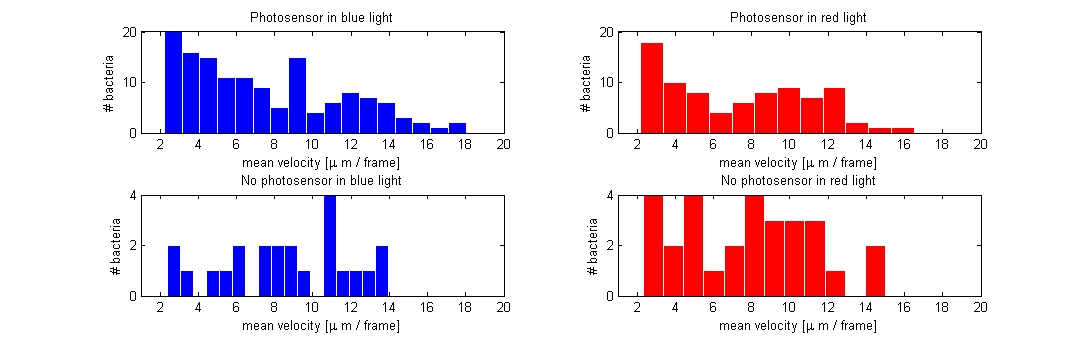
| - | We started out with extracting the mean velocity in | + | We started out with extracting the mean velocity in µm/frame for the photosensor and wildtype bacteria when exposed to red and blue light, the mean was extracted from around 100 paths for each of the 4 different samples (only taken from the experiment where we shifted between red and blue light. Data from the light intensity and gradient experiment were not included). Based on the assumption that bacteria that tumble less would have a higher mean velocity than of those exhibiting a normal rate of tumbling, we would expect a difference in mean velocity of the bacteria expressing the photosensor when illuminated with either red or blue light.<br> |
| - | [[Image:Red_blue_PS_noPS.jpg|750px|center]]<br> | + | [[Image:Red_blue_PS_noPS.jpg|750px|center|]]<br> |
| - | Contrary to our expectations the plot | + | Contrary to our expectations the plot of mean velocity per frame did not show a noticeable difference between the four samples. The photosensor containing bacteria had about the same velocity/frame in blue and red light and there was no clear difference when compared to the wildtype either. The reason for this could be that we were not effective enough at excluding non-informative paths from the data file (i.e. bacteria that are trapped in a circular motion or bacteria only exhibiting Brownian motion) and / or that the conditions for which the experiments were carried out, were not optimal given that the photosensor containing bacteria were exposed to light, before the measurements.<br> |
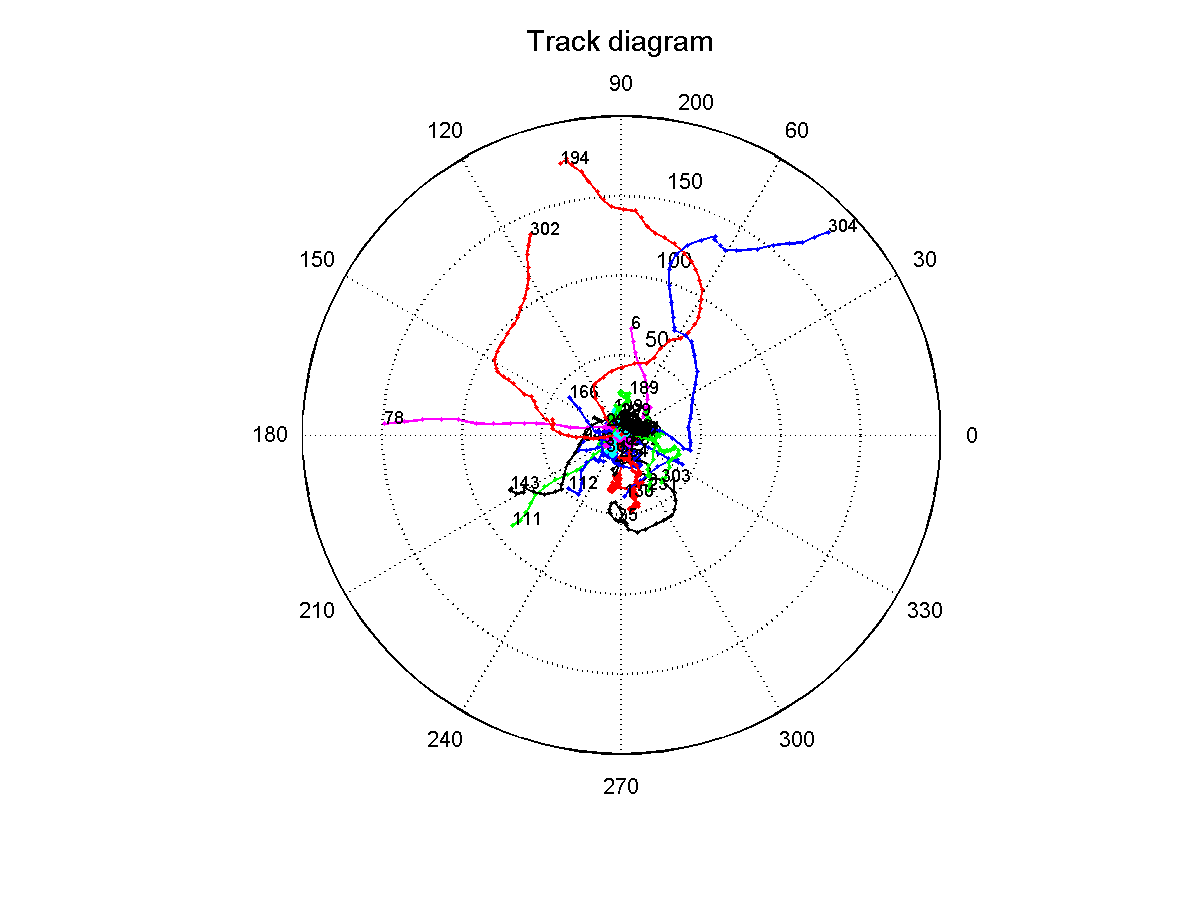
| - | Since the results from the experiment with the red and blue light were inconclusive, we went on to analyse the data from the experiment, where we set up a light gradient in the area that was observed through the microscope. We recorded this for 40 seconds with a framerate of 4 frames/second, waited for 100 seconds and recorded again for 40 seconds. We did 10 iterations of this, which would give us a long-term overview of migration (if there is any) along the light gradient. Afterwards we repeated the same experiment without stimulating the bacteria through a light gradient. A track diagram was created, which should show us if the tracked microbes would show any preference in one direction. In the circular diagram each bacteria starts at point zero and then moves | + | Since the results from the experiment with the red and blue light were inconclusive, we went on to analyse the data from the experiment, where we set up a light gradient in the area that was observed through the microscope. We recorded this for 40 seconds with a framerate of 4 frames/second, waited for 100 seconds and recorded again for 40 seconds. We did 10 iterations of this, which would give us a long-term overview of migration (if there is any) along the light gradient. Afterwards we repeated the same experiment without stimulating the bacteria through a light gradient. A track diagram was created, which should show us if the tracked microbes would show any preference in one direction. In the circular diagram each bacteria starts at point zero and then moves as plotted by its angle and distance traveled. This will give us an idea if there is any tendency to either move towards or away from the light. The lightsource was placed at 90 degrees in the diagram, which means that bacteria that moved in that general direction moved towards the source of light: |
| - | [[Image:TrackDiagramGradient.png|thumb|400px|center|Track diagram of phototaxic bacteria swimming, when exposed to a blue light gradient (470nm). ]]<br> | + | [[Image:TrackDiagramGradient.png|thumb|400px|center|thumb|'''Figure 7:''' Track diagram of phototaxic bacteria swimming, when exposed to a blue light gradient (470nm).]]<br> |
| - | The diagram shows that the bacteria that exhibit proper swimming motility (i.e. paths longer than 40, no circular movement, no | + | The diagram shows that the bacteria that exhibit proper swimming motility (i.e. paths longer than 40, no circular movement, no Brownian movement) show a slight tendency to move towards the more illuminated areas of the gradient. This can be interpreted as blue light acting as an attractant on the phototaxis pathway. Even though we can see this tendency, it should only be used as an indicator, since due to time constraints this experiment could not be repeated and the number of sampled bacteria is rather low (around 40), which means that the shown tendency could also be coincidence.<br> |
The data from the experiments with varying light intensity could not be analysed due to lack of time, but if there is any interest in it, we will gladly provide all the data that we collected.<br> | The data from the experiments with varying light intensity could not be analysed due to lack of time, but if there is any interest in it, we will gladly provide all the data that we collected.<br> | ||
| - | From these experiments, we cannot definitely conclude that blue light acts as an attractant stimulus on part K343007, but the | + | From these experiments, we cannot definitely conclude that blue light acts as an attractant stimulus on part K343007, but the experiments all together indicate that this might be the case. <br> In order to verify if the shown tendency would still show up with a greater pool of samples, dublicate determinations of the light gradient experiment would have to be made. The data analysis also needs more time, since our team could only spend one day doing the experiments and another two days analyzing the results. So there are still a lot of parameters, like tumbling frequency, that simply have not been calculated. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
=== Stability assay === | === Stability assay === | ||
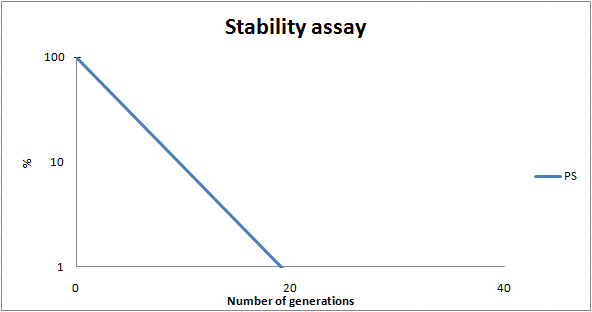
| - | To determine the stability of our pSB1C3-K343007 plasmid, a stability experiment was carried out according to protocol[[https://2010.igem.org/Team:SDU-Denmark/protocols#Stability_assay SA1.1]]. ''E.coli'' MG1655/pSB1C3-K343007 was grown in LB media without chloramphenicol, whereby no selection pressure is exerted on the bacteria. Dilutions of the culture was spread on LA plates and LA plates with | + | To determine the stability of our pSB1C3-K343007 plasmid, a stability experiment was carried out according to protocol[[https://2010.igem.org/Team:SDU-Denmark/protocols#Stability_assay SA1.1]]. ''E.coli'' MG1655/pSB1C3-K343007 was grown in LB media without chloramphenicol, whereby no selection pressure is exerted on the bacteria. Dilutions of the culture was spread on LA plates and LA plates with 35µg/mL chloramphenicol, respectively, and the colony forming units (cfu) was determined for each plate. The CFU for the LA plates represents the total amount of bacteria in the culture, and the CFU of LA plates with chloramphenicol correspond to the amount of resistant bacteria (those carrying the plasmid). The percentage of the total amount of bacteria carrying the plasmid was plotted in a semi-logarithmic graph as a function of number of generations.<br><br> |
| - | [[Image:Team-SDU-PS stab. 2.png|thumb|center|400px|Stability curve of ''E. coli'' strain MG1655-pSB1C3-K343007 showing that the bacteria shed the plasmids within 20 generations. All data can be seen under [https://static.igem.org/mediawiki/2010/8/88/Team-SDU-_PS_atability_assay.ZIP Raw data]]] | + | [[Image:Team-SDU-PS stab. 2.png|thumb|center|400px|'''Figure 8:''' Stability curve of ''E. coli'' strain MG1655-pSB1C3-K343007 showing that the bacteria shed the plasmids within 20 generations. All data can be seen under [https://static.igem.org/mediawiki/2010/8/88/Team-SDU-_PS_atability_assay.ZIP Raw data]]] |
<br><br> | <br><br> | ||
As seen in the graph, almost all of the bacteria had shed the plasmid after 20 generations, suggesting that the plasmid is only stable within the cell for a few generations (<20). This is presumably due to the strain brought upon the bacteria by the plasmid. Thereby when the bacteria are carrying a high-copy plasmid like pSB1C3-K343007 it is plausible that the bacteria will quickly shed the plasmid when no longer exposed to a selection pressure. | As seen in the graph, almost all of the bacteria had shed the plasmid after 20 generations, suggesting that the plasmid is only stable within the cell for a few generations (<20). This is presumably due to the strain brought upon the bacteria by the plasmid. Thereby when the bacteria are carrying a high-copy plasmid like pSB1C3-K343007 it is plausible that the bacteria will quickly shed the plasmid when no longer exposed to a selection pressure. | ||
| Line 84: | Line 79: | ||
=== Growth assay === | === Growth assay === | ||
| - | The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at | + | The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at 550nm every hour for 12 hours and at hour 24. In the experimental setup we used, no lag phase was observed in any of the measurements. <br> The graph below shows the growth of our wild type ''E. coli'' strain MG1655, the MG1655/pSB3T5-K343007 and MG1655/pSB1C3-K343007 respectively. <br><br> |
| - | [[Image:Team SDU-Denmark OD WT+PS.JPG|thumb|center|400px|Growth curve showing measured OD at 550nm. Samples were taken every hour over a 12 hour perion + one sample after 24 hours. In graph ''E. coli'' strain MG1655-pSB3T5-k343007 and MG1655-pSB1C3-K343007 is compaired with Wild type ''E. coli'' strain MG1655 showing no significant difference between the three graphs. No Lag phase is seen. All data can be seen under [https://static.igem.org/mediawiki/2010/2/21/Team_SDU_Denmark_Growth_rate_assay_2_PS.zip Raw data] ]]<br><br> | + | [[Image:Team SDU-Denmark OD WT+PS.JPG|thumb|center|400px|'''Figure 9:'''Growth curve showing measured OD at 550nm. Samples were taken every hour over a 12 hour perion + one sample after 24 hours. In graph ''E. coli'' strain MG1655-pSB3T5-k343007 and MG1655-pSB1C3-K343007 is compaired with Wild type ''E. coli'' strain MG1655 showing no significant difference between the three graphs. No Lag phase is seen. All data can be seen under [https://static.igem.org/mediawiki/2010/2/21/Team_SDU_Denmark_Growth_rate_assay_2_PS.zip Raw data] ]]<br><br> |
| - | From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite | + | From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite contradictory to our results obtained from the stability assay. The transitory stability of pSB1C3-K343007 suggests that it is highly unfavorable for the bacteria, wherefore it might be expected that the growth of the bacteria containing this plasmid would be affected. Thus, however much a disadvantage the plasmid pose to the bacteria, their growth are not significantly influenced by the plasmid. The added reproduction load due to the plasmids, might also prolong the lag phase of the bacteria. Whether this is the case can not be concluded based on this experiment as no lag phase was observed in this experiment.<br><br> |
| + | |||
= References = | = References = | ||
Latest revision as of 00:59, 28 October 2010
 "
"