Team:BCCS-Bristol/Wetlab/Achievements
From 2010.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:BCCS-Bristol/Header}} | {{:Team:BCCS-Bristol/Header}} | ||
| - | <center> • [[:Team:BCCS-Bristol/Wetlab/Achievements|Achievements]] • [[ | + | <center> • [[:Team:BCCS-Bristol/Wetlab/Achievements|Achievements]] • [[Team:BCCS-Bristol/Wetlab/Part_Design/BioBricks/PyeaR|Our BioBrick]] • [[:Team:BCCS-Bristol/Wetlab/Beads|Beads]]• [[:Team:BCCS-Bristol/Wetlab/Experiments|Lab Work]] |
| - | [[:Team:BCCS-Bristol/Wetlab/ | + | • [[:Team:BCCS-Bristol/Wetlab/Improvements|Improvements]] • [[:Team:BCCS-Bristol/Wetlab/Safety|Safety]] • </center> |
=Achievements= | =Achievements= | ||
| - | + | {{:Team:BCCS-Bristol/newtoc}} | |
| - | + | Despite having an extremely small wet lab team (only two members) we believe we've achieved a lot this year. We've come from our initial ideas stage in July, to possessing a fully functional prototype product, good enough to catch commercial attention by the end of October. Alongside this, we've also helped better characterise a pre-existing BioBrick. | |
| - | + | ||
| - | Despite having an extremely small wet lab team (only two members) we believe we've achieved a lot this year. We've come from our initial ideas stage in July, to possessing a fully functional prototype product, good enough to catch commercial attention by the end of October. Alongside this, we've also helped better characterise a pre-existing | + | |
Below is a brief summary of our biggest achievements | Below is a brief summary of our biggest achievements | ||
| - | + | ==A Well Characterised New BioBrick== | |
| - | ==A Well Characterised New | + | |
| Line 21: | Line 18: | ||
| - | The focal point of our wetlab work was the construction and characterisation of a new | + | The focal point of our wetlab work was the construction and characterisation of a new BioBrick [http://partsregistry.org/Part:BBa_K381001 BBa_K381001]. This part is a composite of two pre-existing BioBricks, with details of its design available [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/Part_Design/BioBricks/PyeaR| here]. |
| Line 28: | Line 25: | ||
We're proud of this part, and hope it will be useful to other teams in the future. | We're proud of this part, and hope it will be useful to other teams in the future. | ||
| - | + | <br/><br/><br/><br/><br/><br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Elegant Solution to Signal Calibration== | ==Elegant Solution to Signal Calibration== | ||
| Line 40: | Line 33: | ||
| - | As well as making our | + | As well as making our BioBrick, we also worked further to develop a product that would actually function. As part of this we altered the traditional signal detection method of measuring output to remove the problem of misleading figures. Our new method of instead detecting the ratio of two outputs removes difficulties in distinguishing between large signals due to high Nitrate levels and those due simply to large colonies. |
| Line 46: | Line 39: | ||
| - | For more information on how the ratio method works and our results from it, click [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/characterising_ratio | + | For more information on how the ratio method works and our results from it, click [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/characterising_ratio here] |
==Novel Use of Cell Encapsulation== | ==Novel Use of Cell Encapsulation== | ||
| Line 61: | Line 54: | ||
For more information on how the beads work and how they can be made, click [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/Beads| here]. We hope the resources and data we've provided will encourage other iGEM teams to pursue this method in the future. | For more information on how the beads work and how they can be made, click [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/Beads| here]. We hope the resources and data we've provided will encourage other iGEM teams to pursue this method in the future. | ||
| - | ==Characterising A Pre-Existing | + | |
| + | ==Characterising A Pre-Existing BioBrick== | ||
| Line 67: | Line 61: | ||
| - | As well as developing our own | + | As well as developing our own BioBrick, we also worked to better characterise one already in the parts registry [http://partsregistry.org/Part:BBa_K216005| BBa_K216005]. Graph 3 shows just one of many Miller assays we conducted to analyse its performance, more of which can be found [https://2010.igem.org/Team:BCCS-Bristol/Wetlab/BetaGalactosidaseAssays here]. |
Latest revision as of 23:56, 27 October 2010
iGEM 2010
Achievements
Contents |
Below is a brief summary of our biggest achievements
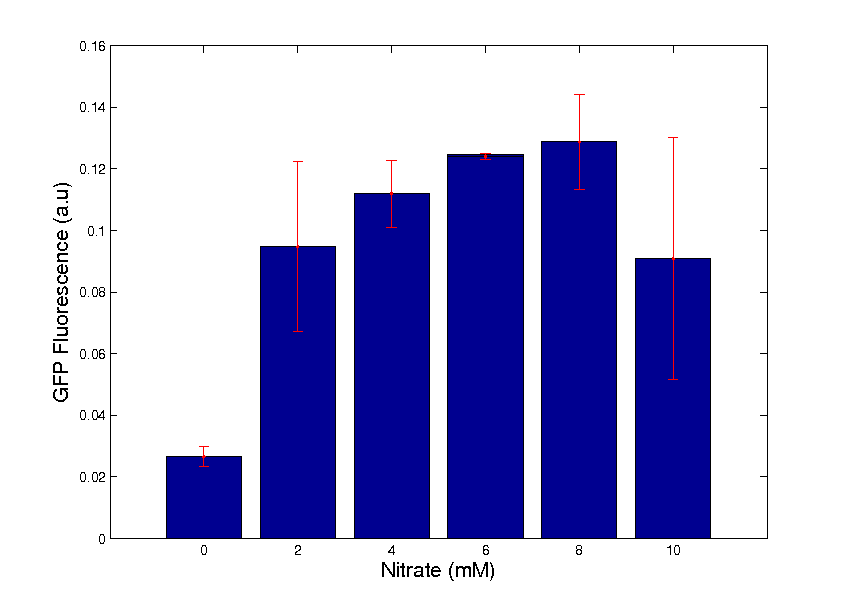
A Well Characterised New BioBrick
The focal point of our wetlab work was the construction and characterisation of a new BioBrick [http://partsregistry.org/Part:BBa_K381001 BBa_K381001]. This part is a composite of two pre-existing BioBricks, with details of its design available here.
As shown in graph 1, we mapped its behaviour across the range where it was known to be most sensitive, further details of which can be found here.
We're proud of this part, and hope it will be useful to other teams in the future.
Elegant Solution to Signal Calibration
As well as making our BioBrick, we also worked further to develop a product that would actually function. As part of this we altered the traditional signal detection method of measuring output to remove the problem of misleading figures. Our new method of instead detecting the ratio of two outputs removes difficulties in distinguishing between large signals due to high Nitrate levels and those due simply to large colonies.
As is evident in graph 2, we well characterised this method and found it to work well, particularly within the range relevant to our project. Our results were even good enough to allow the construction of a formula linking signals detected to levels in the soil.
For more information on how the ratio method works and our results from it, click here
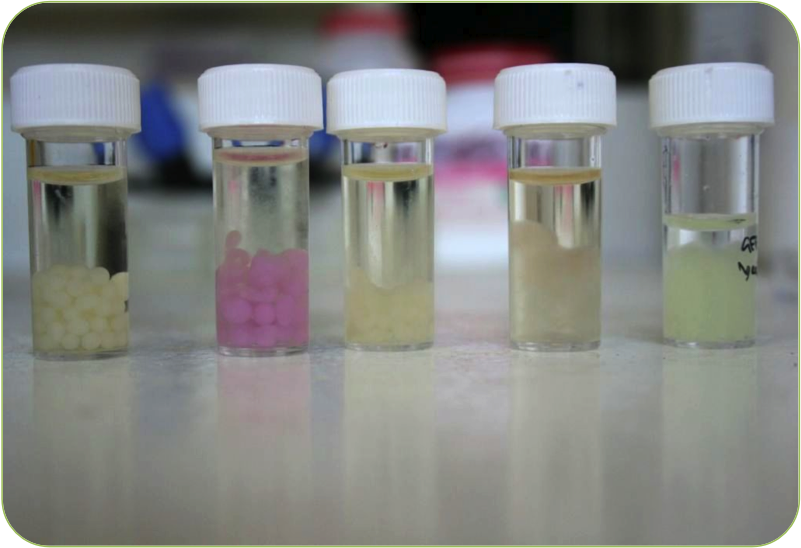
Novel Use of Cell Encapsulation
In what we believe is a first in iGEM, we used technology to both allow safe use of our E. coli in the public and environment dramatically improve signal detection.
Our Bead encapsulation method ensures a stronger signal that can actually be detected by our end user (farmers) without high tech equipment. It also reduces the risk of our bacteria escaping unchecked in the environment, and assisted in our human practices side of the project; making our product more appealing to the public.
For more information on how the beads work and how they can be made, click here. We hope the resources and data we've provided will encourage other iGEM teams to pursue this method in the future.
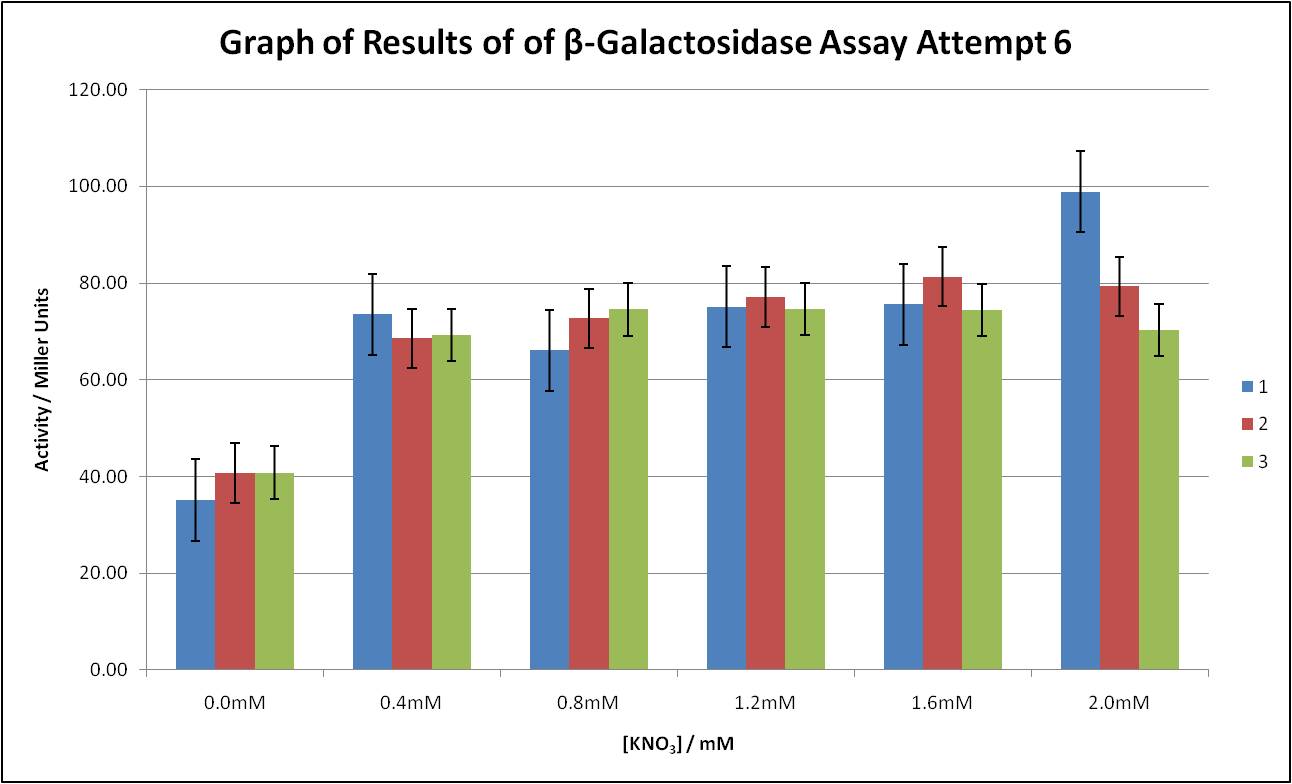
Characterising A Pre-Existing BioBrick
As well as developing our own BioBrick, we also worked to better characterise one already in the parts registry [http://partsregistry.org/Part:BBa_K216005| BBa_K216005]. Graph 3 shows just one of many Miller assays we conducted to analyse its performance, more of which can be found here.
 "
"