Team:TU Munich/Project
From 2010.igem.org
(→in silico design of switching and trigger unit) |
Hartlmueller (Talk | contribs) (→Network construction) |
||
| (93 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
= Vision= | = Vision= | ||
| - | Until today, 13.628 biobrick sequences<sup>[[Team:TU_Munich/Project#ref1|[1]]]</sup> have been submitted to partsregistry, thereof 102 reporter units | + | Until today, 13.628 biobrick sequences<sup>[[Team:TU_Munich/Project#ref1|[1]]]</sup> have been submitted to partsregistry, thereof 102 reporter units and 12 signaling bricks. |
| - | Since | + | Since then, people are trying to arrange these single biological building blocks in such a manner that allows producing special biotechnological products (metabolic engineering), developing biological sensory circuits (biosensors) and even giving microorganisms the ability to react on multiple environmental factors and serve both as disease indicator and drug. These examples and further promising ideas were implemented on previous iGEM-competitions.<sup>[[Team:TU_Munich/Project#ref2|[2]]]</sup><sup>[[Team:TU_Munich/Project#ref3|[3]]]</sup><sup>[[Team:TU_Munich/Project#ref4|[4]]]</sup> <br><br> |
| - | The idea of combining the outcome of several iGEM competitions to construct complex synthetic biological systems falls at the last hurdle - the fact, that each team uses a different principle how to access and functionally connect the respectively used biobricks. For example, it is a major challenge to create a system that uses several sensoring BioBricks from different iGEM-teams which in turn regulates reportering BioBricks from various teams. In order to combine and fully take advantage of these promising projects, our vision is to develop an adapter that allows interconnecting | + | The idea of combining the outcome of several iGEM competitions to construct complex synthetic biological systems falls at the last hurdle - the fact, that each team uses a different principle how to access and functionally connect the respectively used biobricks. For example, it is a major challenge to create a system that uses several sensoring BioBricks from different iGEM-teams which in turn regulates reportering BioBricks from various teams. In order to combine and fully take advantage of these promising projects, our vision is to develop an adapter that allows interconnecting arbitrary biobricks on a functional level. Such a system easily allows to setup sensor-reporter circuits and interconnect them to complete biological chips... A further step towards artificial cells.<br><br> |
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | Generally speaking, the above | + | Generally speaking, the above adapter has to meet the following requirements: |
*'''Universality''' | *'''Universality''' | ||
:The adapter has to be compatible to as many BioBricks as possible. This objective will guarantee that a large number of BioBricks can be connected. | :The adapter has to be compatible to as many BioBricks as possible. This objective will guarantee that a large number of BioBricks can be connected. | ||
| Line 17: | Line 17: | ||
:Once the basic design of the system is established, the construction of the system is supposed to be automated in silico. This way it will be possible to create an adapter connecting a large amount of BioBricks. | :Once the basic design of the system is established, the construction of the system is supposed to be automated in silico. This way it will be possible to create an adapter connecting a large amount of BioBricks. | ||
*'''Biological orthogonality''' | *'''Biological orthogonality''' | ||
| - | : | + | :Interference with cellular components has to be as low as possible in order to avoid unwanted and perturbing side effects. |
*'''Logic''' | *'''Logic''' | ||
:The adapter is supposed to not only associate different BioBricks, but to functionally connect BioBricks in a precisely determined manner (including operations such as AND/OR/NOT). | :The adapter is supposed to not only associate different BioBricks, but to functionally connect BioBricks in a precisely determined manner (including operations such as AND/OR/NOT). | ||
<br> | <br> | ||
| - | Several biological logic units, devices and circuits have been developed so far<sup>[[Team:TU_Munich/Project#ref5|[5]]]</sup>, but to our knowledge, none was shown to meet all | + | Several biological logic units, devices and circuits have been developed so far<sup>[[Team:TU_Munich/Project#ref5|[5]]]</sup>, but to our knowledge, none was shown to meet all requirements listed above. |
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| + | |||
=Implementation= | =Implementation= | ||
To functionally connect BioBricks, there are several possibilities including genetic switches, riboswitches and direct protein-protein interactions. We investigated several hypothetically principles, and decided to focus our practical work on the development of a RNA-RNA interaction-based switch. These switches are capable of changing between two states, a state of antitermination and termination, and make use of highly-specific RNA-RNA interaction. In principle such a switch can fulfill all requirements mentioned previously. The following text clarifies how these switches work in detail. | To functionally connect BioBricks, there are several possibilities including genetic switches, riboswitches and direct protein-protein interactions. We investigated several hypothetically principles, and decided to focus our practical work on the development of a RNA-RNA interaction-based switch. These switches are capable of changing between two states, a state of antitermination and termination, and make use of highly-specific RNA-RNA interaction. In principle such a switch can fulfill all requirements mentioned previously. The following text clarifies how these switches work in detail. | ||
| Line 31: | Line 32: | ||
{| | {| | ||
|- | |- | ||
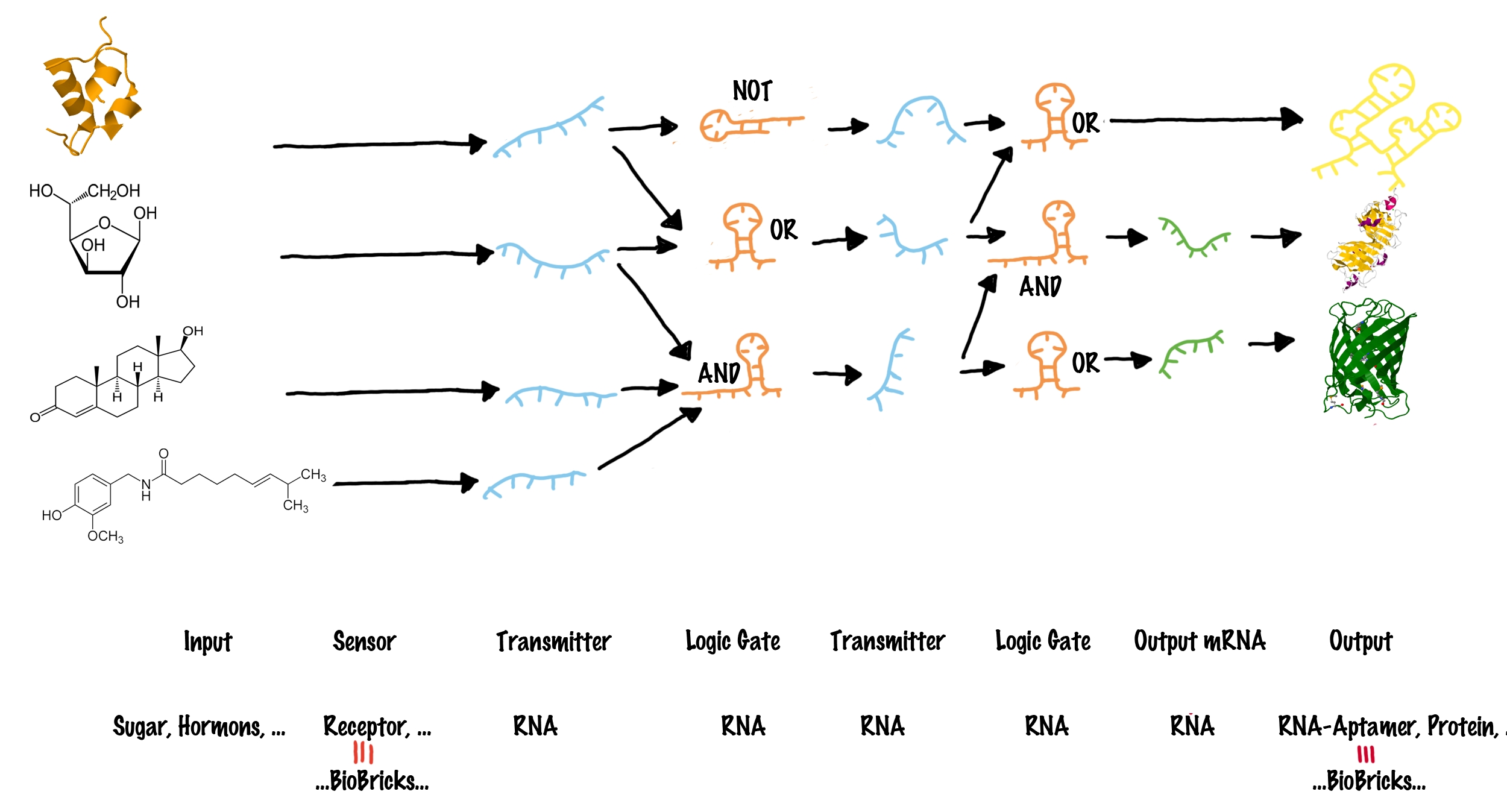
| - | |[[Image:Networks.png|center| | + | |[[Image:Networks.png|center|thumb|730px|The general principle how different inputs can be connect to various outputs. For details see text.<br>Inputs (such as proteins or small molecules) are indicated on the left side. blue lines represent transmitter molecules whereas organe lines present logic gates. The type of logic gate is indicated. Green lines indicate transmitter RNA that can function as mRNA and consequently generate any output gene (indicated on the very right).]] |
|} | |} | ||
In order to connect different BioBricks, our network requires four major types of components: | In order to connect different BioBricks, our network requires four major types of components: | ||
| Line 40: | Line 41: | ||
{{:Team:TU_Munich/Templates/InfoBoxStart}}'''Computer vs. molecular network - and our approach'''<br> | {{:Team:TU_Munich/Templates/InfoBoxStart}}'''Computer vs. molecular network - and our approach'''<br> | ||
| - | Logic gates in a molecular network are often compared to transistors used in a computer, where billions of transistors are incorporated<sup>[[Team:TU_Munich/Project#ref7|[7]]]</sup>. The main advantage on a computer chip is, all transistors share the same functional principle, and only the way connecting them in a special sequence allows specific addressing of only a subset of other transistors by an input. However, | + | Logic gates in a molecular network are often compared to transistors used in a computer, where billions of transistors are incorporated<sup>[[Team:TU_Munich/Project#ref7|[7]]]</sup>. The main advantage on a computer chip is, all transistors share the same functional principle, and only the way connecting them in a special sequence allows specific addressing of only a subset of other transistors by an input. However, spatially fixed connections of molecular logic gates are not possible in a living cell. The "wiring" within a cell relies on the specific interaction between transmitter molecule and their corresponding logic gates, for example implemented by protein-protein/ligand-protein interactions or specific ligand-riboswitch interactions.<sup>[[Team:TU_Munich/Project#ref8|[8]]]</sup><sup>[[Team:TU_Munich/Project#ref9|[9]]]</sup> As a result, in a cell, each occurring logic gate ("transistor") has to be different, at least in a special recognition site<sup>[[Team:TU_Munich/Project#ref10|[10]]]</sup> - for example like different transcription factors, recognizing different DNA-sites. Thanks to evolution, nature easily can invent a new transistor for each task - science achieves this only on a limited scale, and producing synthetic molecular logic gates artificially by either rational or evolutionary protein or riboswitch engineering, is limited to small circuits so far<sup>[[Team:TU_Munich/Project#ref11|[11]]]</sup>. Our project aims to establish a molecular switch as close as possible to a electronic transistor, thus sharing the same functional principle for all logic gates. At the same time, we want to design a easily exchangeable recognition site, which can individually be designed by everyone! {{:Team:TU_Munich/Templates/InfoBoxEnd}} |
| - | These elements can be combined to build up a molecular network (see illustration). Each input molecule (such as a BioBrick) produces a unique transmitter molecule. All transmitters belong to the same type of molecule and share a common design. However each transmitter molecule can only interact and activate a certain subset of logic gates. In other words, logic gates have to recognize as well as bind the corresponding transmitter molecules and are capable of producing a new output transmitter molecule. Depending on the type of the logic gate (AND, OR or NOT<sup>[[Team:TU_Munich/Project#ref6|[6]]]</sup>), an output transmitter is only created if both input transmitter molecules are present (AND), at least one of two input transmitters is present (OR) or if no input transmitter is present at all (NOT). Once a logic | + | These elements can be combined to build up a molecular network (see illustration). Each input molecule (such as a BioBrick) produces a unique transmitter molecule. All transmitters belong to the same type of molecule and share a common design. However, each transmitter molecule can only interact and activate a certain subset of logic gates. In other words, logic gates have to recognize as well as bind the corresponding transmitter molecules and are capable of producing a new output transmitter molecule. Depending on the type of the logic gate (AND, OR or NOT<sup>[[Team:TU_Munich/Project#ref6|[6]]]</sup>), an output transmitter is only created if both input transmitter molecules are present (AND), at least one of two input transmitters is present (OR) or if no input transmitter is present at all (NOT). Once a logic gate has produced a new output transmitter, these transmitters can in turn address another subset ("layer") of logic gates. In theory many layers of logic gates can be connected this way allowing the creation of large networks. Until this step, various transmitter molecules might have been produced. But in order to create a Biobrick output, the last layer of logic gates finally generates transmitter molecules that will not active logic gates, but will rather interact with the cell metabolism to produce a BioBrick response. In other words, the last layer of transmitter molecules is capable of regulating BioBrick formation. |
| - | Summarizing, the network | + | Summarizing, the network establishes a connection between input BioBricks and output BioBricks in a functional manner. |
Having addressed the basic layout of the molecular network, the next step is to determine what type of molecules can perform the required functions. We decided to use RNA, both as transmitter molecules and for constructing logic gates. Several advantages result from the utilization of RNA as the central element: | Having addressed the basic layout of the molecular network, the next step is to determine what type of molecules can perform the required functions. We decided to use RNA, both as transmitter molecules and for constructing logic gates. Several advantages result from the utilization of RNA as the central element: | ||
*During the last years, many Biobricks were designed that are sensitive to various chemicals and substances. These BioBricks often function as a transcription factor that binds to a specific DNA sequence and consequently would be capable to produce a specific transmitter RNA molecule. Thus, in principle each BioBrick which involves transcription can be integrated in our network. | *During the last years, many Biobricks were designed that are sensitive to various chemicals and substances. These BioBricks often function as a transcription factor that binds to a specific DNA sequence and consequently would be capable to produce a specific transmitter RNA molecule. Thus, in principle each BioBrick which involves transcription can be integrated in our network. | ||
| Line 56: | Line 57: | ||
==Design and functional principle of logic gates== | ==Design and functional principle of logic gates== | ||
The concept introduced above provides a framework that can potentially serve as an universal adapter between different BioBricks. However, the [[Team:TU_Munich/Glossary#logic gate | logic gates]] have not been specified more precisely so far. This will be done in the following section. | The concept introduced above provides a framework that can potentially serve as an universal adapter between different BioBricks. However, the [[Team:TU_Munich/Glossary#logic gate | logic gates]] have not been specified more precisely so far. This will be done in the following section. | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | Generally speaking, our logic gates are to | + | Generally speaking, our logic gates are to possess the following characteristics: |
*Logic gates, such as AND, OR and NOT, have to be implemented by RNA-interaction based principles (see [[Team:TU_Munich/Project#How_to_connect_BioBricks | How to connect BioBricks]]). | *Logic gates, such as AND, OR and NOT, have to be implemented by RNA-interaction based principles (see [[Team:TU_Munich/Project#How_to_connect_BioBricks | How to connect BioBricks]]). | ||
| - | *All logic gates have to recognize their corresponding [[Team:TU_Munich/Glossary#Transmitter (bioLOGICS)| transmitter | + | *All logic gates have to recognize their corresponding [[Team:TU_Munich/Glossary#Transmitter (bioLOGICS)| transmitter RNAs]] and, in response, produce an output transmitter molecule. |
*Logic gates should follow a basic design rule, in such a way, that their creation can be automated ''in silico''. | *Logic gates should follow a basic design rule, in such a way, that their creation can be automated ''in silico''. | ||
*The response efficiency of a logic gate toward a transmitter molecule should be comparable for all logic gates to provide calculable robustness and sensitivity. This will ensure comparable molecular concentrations and functionality of large networks. | *The response efficiency of a logic gate toward a transmitter molecule should be comparable for all logic gates to provide calculable robustness and sensitivity. This will ensure comparable molecular concentrations and functionality of large networks. | ||
| - | *The system has to be designed for ''in vivo'' utilization at the first place. As a reference we always assumed a temperature of 37 °C and an E. coli environment. | + | *The system has to be designed for ''in vivo'' utilization at the first place. As a reference we always assumed a temperature of 37 °C and an ''E. coli'' environment. |
| - | + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | |
| - | In order to build | + | In order to build logic gates for our bioLOGICS system we will first create a simple switch. A switch can be activated by one transmitter RNA and produce an output transmitter RNA. In contrast to a logic gate, a switch does not perform logic operations. However by combining switches, logic gates can be created. The following text will first describe how the developed switch works and secondly, how logic gates such as AND/OR/NOT can be created using these switches. |
| - | < | + | <html> |
| - | <br> | + | <script type="text/javascript"> |
| - | === | + | $(document).ready(function(){ |
| - | Roughly speaking, a | + | |
| + | $("p.trigger").click(function(){ | ||
| + | $(this).next("div").find(".thumbinner").slideToggle("slow"); | ||
| + | |||
| + | return false; //Prevent the browser jump to the link anchor | ||
| + | }); | ||
| + | $("a.toggle_close").click(function(){ | ||
| + | var nextParent = $(this).parent(); | ||
| + | while(nextParent!= null) { | ||
| + | if(!nextParent.is(".toggle_container")) { | ||
| + | nextParent = nextParent.parent(); | ||
| + | } | ||
| + | else { | ||
| + | break; | ||
| + | } | ||
| + | } | ||
| + | if(nextParent.is(".toggle_container")) { | ||
| + | |||
| + | |||
| + | nextParent.prev("div").find(".thumbinner").slideToggle("slow"); | ||
| + | |||
| + | |||
| + | } | ||
| + | |||
| + | |||
| + | return false; //Prevent the browser jump to the link anchor | ||
| + | }); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart1}}Read more{{:Team:TU Munich/Templates/ToggleBoxStart2}} | ||
| + | [[Image:toggle_switch.png|500px|thumb|center|id="hideOnReadMore"|'''A''' The basic structure of a bioLOGICS switch (left) and a transmitter molecule (right).<br>'''B'''The process of switching. See the text in the close-by "Read more" section for details.<br>Rectangles present the composition of our functional units on the level of DNA. Fringed lines represent RNA produced by RNA polymerase. The stem loop structure depicts the switchable terminator. Terminator and target site are illustrated in blue and turquoise, respectively. Recognition sites are indicated in different colors, in this case red for the input transmitter and green for the output transmitter.Each switch and or later logical unit has to be flanked by a promotor and another constitutive terminator, to allow RNA-production by RNA-polymerase in a proper way. ]] | ||
| + | {{:Team:TU Munich/Templates/ToggleBoxStart3}} | ||
| + | ===Switch=== | ||
| + | [[Image:TUM2010_switch-and-transmitter.jpg|550px|right|thumb|The basic strcutrue of a switch (left) and a transmitter RNA (right). See text for details.]] | ||
| + | Roughly speaking, a switch can be regarded as an enhanced switchable transcriptional terminator. The enhancement can be described easier by dividing a switch into its functional components: | ||
*'''Target site'''<br> | *'''Target site'''<br> | ||
:The target site is the functional core element of our switches, allowing a shift between an "on" and "off" state. Since we work on the level of RNA-production (transcription), a "switchable" transcriptional terminator is suitable for this purpose. By allowing or preventing formation of a transcriptional terminator, that is by switching between termination and antitermination it is possible to represent an "off" and an "on" state, respectively. Therefore, the target site is the 5' ending of the terminator and is required for a stable terminator formation. It should be noted that this principle was also observed in nature. | :The target site is the functional core element of our switches, allowing a shift between an "on" and "off" state. Since we work on the level of RNA-production (transcription), a "switchable" transcriptional terminator is suitable for this purpose. By allowing or preventing formation of a transcriptional terminator, that is by switching between termination and antitermination it is possible to represent an "off" and an "on" state, respectively. Therefore, the target site is the 5' ending of the terminator and is required for a stable terminator formation. It should be noted that this principle was also observed in nature. | ||
:To highlight and illustrate the functional principle of our switches, only the part of the terminator which is involved in interacting with a transmitter molecule and which is responsible for shifting between "on" and "off" state is called target site. The remaining terminator sequence is called terminator in the following, even if both, target site and terminator build up the terminator structure occurring in nature. | :To highlight and illustrate the functional principle of our switches, only the part of the terminator which is involved in interacting with a transmitter molecule and which is responsible for shifting between "on" and "off" state is called target site. The remaining terminator sequence is called terminator in the following, even if both, target site and terminator build up the terminator structure occurring in nature. | ||
| + | :The important aspect of our switches is the fact that all switches will hold the same identical target site. Therefore having found one functional "switchable" terminator, will allow almost unlimited upscaling since this terminator can be used for a large library of switches. This is the main difference to previous works done on this field, which always required developing a new shifting principle for each switch.<sup>[[Team:TU_Munich/Project#ref12|[12]]]</sup><sup>[[Team:TU_Munich/Project#ref13|[13]]]</sup><sup>[[Team:TU_Munich/Project#ref14|[14]]]</sup> Beside this scalability, this principle provides a comparable on/off shifting rate (responds function) for all switches, avoiding complex fine tuning of molecular networks. | ||
| + | :To sum it up, the target site, allows to switch between an "on" and "off" state. But so far, the switch is not capable of performing specific interaction with transmitter molecules. This is where the recognition site comes into play. | ||
| + | *'''Recognition site''' | ||
| + | :The recognition site defines which transmitter molecule can actually interact with the switch. Therefore, a unique recognition site is generated for each switch and is positioned right upstream of the target site. In principle the recognition can be any random sequence as long as it remains unique within the molecular network. | ||
| + | Summing up, the recognition site allows a specific interaction between switches and transmitter molecules. Once this interaction is formed, an interaction between the transmitter and the target will actually switch the state of the terminator. This allows the specific arrangement and interconnection of numerous of these switches by transmitter molecules, without changing the target site. Comparable to wires connecting many identical transistors, our target site remains the same. | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | *''' | + | ===Transmitter RNA´s=== |
| - | : | + | As desccribed above, transmitter RNAs are the input and output of bioLOGICS switches (compare [[Team:TU_Munich/Project#How_to_connect_BioBricks | How to connect BioBricks]]). These transmitters are short ssRNA molecules representing the "trigger" to shift switches between the "on" and "off" state. To fulfill this role, they need to posses the following properties: |
| - | + | *A transmitter may only interact with certain switches. That is, a transmitter has to find the corresponding recognition site of a switch. | |
| - | of | + | *Once an interaction is established between a transmitter and a switch, a transmitter has to be capable of changing the secondary structure of a terminator and thus cause antitermination. |
| - | + | Again, these two properties are fulfilled by two components of the transmitter: | |
| + | *'''Identity site''' | ||
| + | :This site is capable of forcing an interaction between the transmitter and the switch. Therefore it is complementary to the recognition site of this switch. As the recognition site is unique within a network, so is the identity site. However, the single identity site is not capable of changing the state of the switch. That is were the trigger site comes into play. | ||
| + | *'''Trigger site''' | ||
| + | :Once an interaction is created by the identity site, the trigger site is capable of actually shifting the switch since it is complementary to the target site of the switch. To fulfill this role, it is placed upstream at the 5' end of the identity site. As the target site is the same for all switches, the trigger site is the same for all signals. Therefore it is important, that similar to the identity site, a trigger site cannot function on its own. That is, a single trigger site cannot shift the state of a switch without the help of an identity site. | ||
| + | |||
| + | Summing up, we applied the principle introduced for the switches to the transmitter molecules. In contrast to previous approaches on this field <sup>[[Team:TU_Munich/Project#ref12|[12]]]</sup>, we introduced the described synthetic trigger site in such a manner that it is not able to change the state of the terminator on its own, but only in combination with the identity site. So the challenge is to arrange and optimize these elementary building blocks thermodynamically, that a trigger site is only able to switch in combination with its respective identity site. This was done by ''in silico'' design using [[TU Munich/Glossary#NUPACK| NUPACK]], presented in section [[TU Munich/Modeling#in silico design based on thermodynamic calculations| in silico design]]. | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===Putting it all together: the switching process=== | ===Putting it all together: the switching process=== | ||
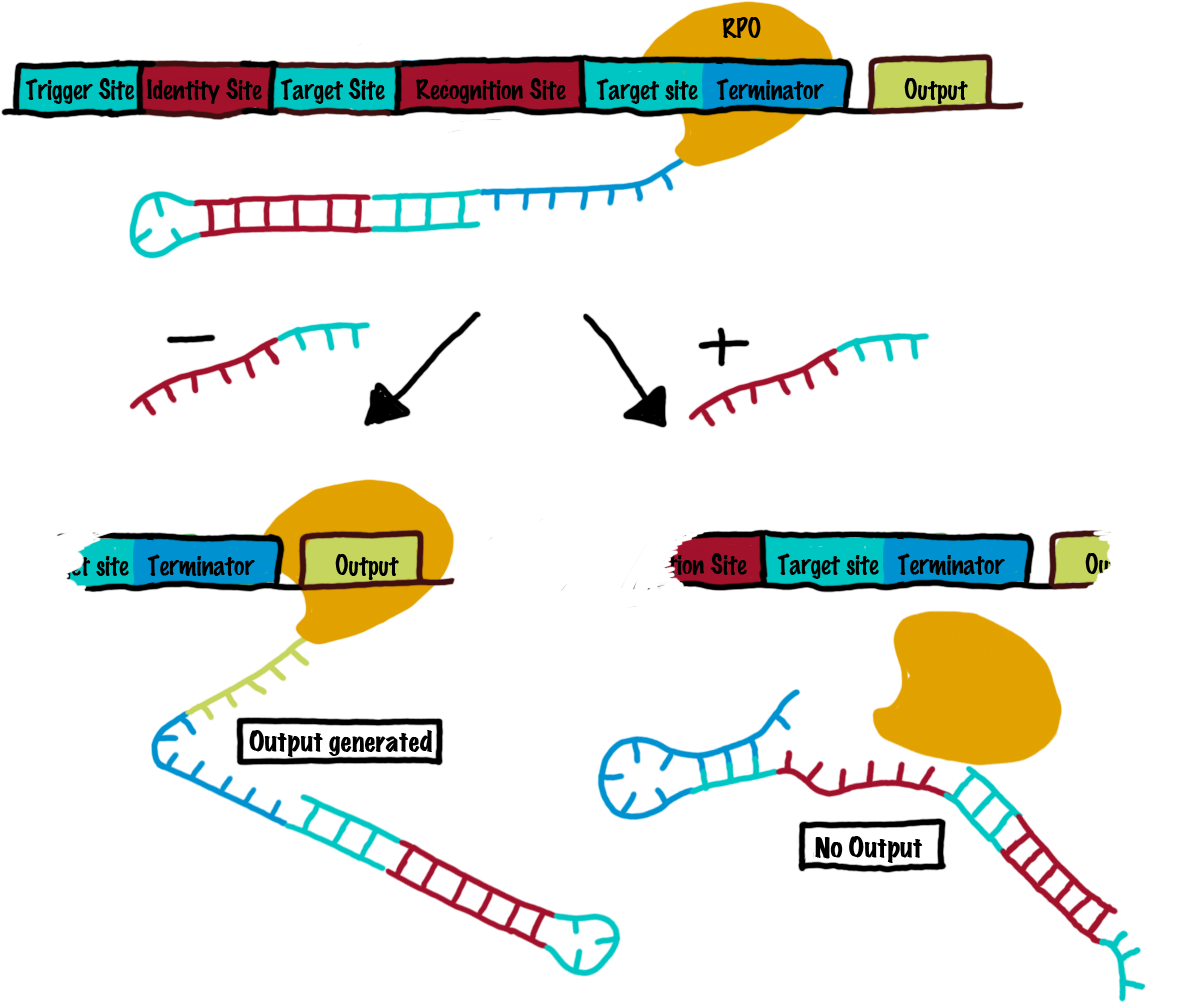
| + | [[Image:TUM2010_switching-process.jpg|550px|right|thumb|The basic structure of a switch (left) and a transmitter RNA (right). See text for details.]]The functional principle of the designed switches is illustrated in the figure. The switch is positioned on DNA upstream of a desired output transmitter. So in the absence of a triggering transmitter molecule, transcription will be canceled by the formation of a RNA stem loop in the nascent RNA-chain. This will cause the RNA polymerase to stop transcription and fall off the DNA and consequently no output RNA will be produced. This process only relies on [[Team:TU_Munich/Glossary#Termination| rho-independent termination]]. | ||
| + | On the other hand, in the presence of a [[Team:TU_Munich/Project#RNA_transmitters | input transmitter]], this small functional RNA inhibits the stem loop formation by complementary base-pairing and hence avoids termination of transcription. In detail, the identity site (red part on transmitter) binds the recognition site (red part on switch) and serves as [[Team:TU_Munich/Glossary#Toehold|toehold]], which will thermodynamically allow the trigger site (turquoise part on transmitter) to perform a [[Team:TU_Munich/Glossary#Strand Displacement| strand displacement]] and open up the stem loop structure. Consequently the polymerase can read all the way through and form the output RNA.<br>Summing up, we use this concept to create a switch that can be toggled by a transmitter RNA molecule and in response, is able to produce another transmitter RNA. | ||
<br> | <br> | ||
| - | |||
| - | |||
<br> | <br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ===From switches towards bioLOGICS logic gates=== | |
| - | === | + | As described, each switch can be accessed by a specific RNA-transmitter molecule, representing the input. In turn, another RNA-transmitter molecule will be produced if the switch shifts its state. This output transmitter of one switch can serve as input transmitter for the next switch by meaningful selection and design of the respective recognition sites. This easily allows arranging several switches in specific sequences and faulty wiring - the corner stone of a logical network.<br> |
| - | As described, each switch can be accessed by a specific RNA-transmitter molecule, | + | To ease the building of logical networks we want to create a switch capable of Boolean logics, a common mathematical principle fundamental for computational science. Since AND/OR/NOT are basic logic operations which can be implemented with the presented switches, all remaining operations (such as XOR, NAND, ...) can be expressed by these three operators according to laws of boolean logics. |
| + | Creating logic gates is achieved by combining two switches in two different ways, as illustrated below. | ||
| + | *AND gate | ||
| + | :An AND gate can be constrcuted by positioning two switches right next to each other. For the output transmitter to be created, both input transmitter have to be present.[[Image:AND2.png|500px|thumb|center|Combining two switches in series creates a logic AND gate.]] | ||
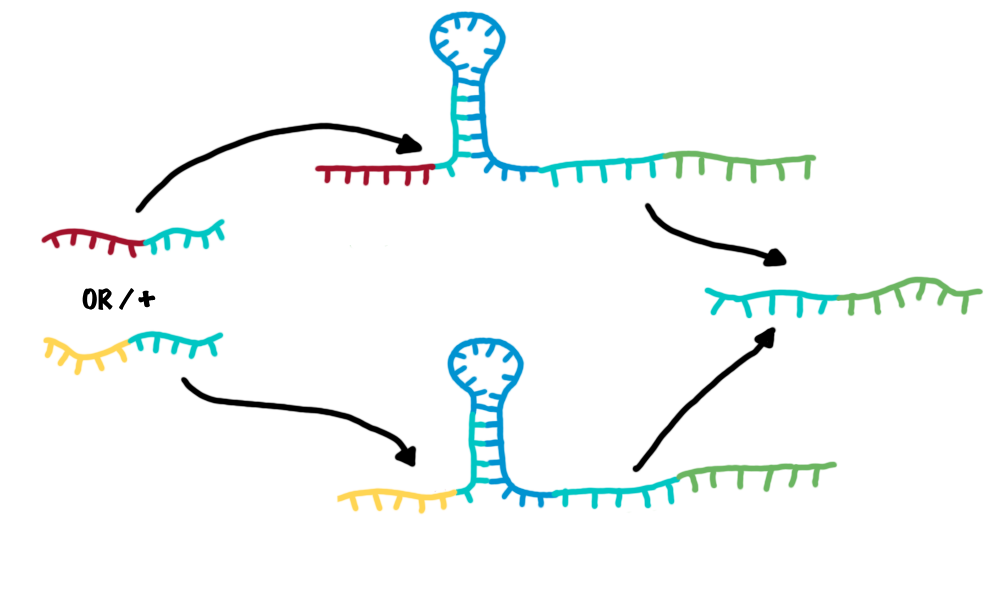
| - | + | *OR gate | |
| - | + | :An OR gate is created by utilizing two independent switches sharing the same output transmitter. If each one of both switches is activated, an output transmitter is generated. Therefore, one input transmitter is enough to produce an output transmitter.[[Image:OR2.png|500px|thumb|center|Combining two switches in parallel creates a logic OR gate.]] | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | *NOT gate | |
| - | + | :A NOT gate is supposed to function as an inverter. In contrast to the gates described above, a not gate requires only one sitch. However, to meet the design rule for transmitter molecules, this switch shows some differences compared to the switches used for AND and OR gates.<br> | |
| - | To | + | :Since the transcriptional terminator may not form if no transmitter is present. Consequently, the switch needs an internal trigger site, capable of preventing terminator formation. To allow the binding of an input transmitter molecule, the switch contains a recognition site upstream of a second target site. The additional target site is mandatory since all transmitter molecules have to carry a trigger. In the case of the NOT switch this trigger site may not bind the actual target site within the transcriptional terminator. In other words, a second target site further upstream is required to catch the trigger site of the transmitter molecule. At the same time, the identity site of the transmitter may not bind right upstream of the terminator. This is accomplished by placing an other identity site right upstream of the terminator rather than an recognition site (compare switches used for AND or OR gate). Due to these two difference, the input transmitter is forced to bind further upstream to the recognition site, displacing the internal trigger site of the switch. This will allow the RNA polymerase to read through an create the output transmitter. |
| + | [[Image:NOT2.png|500px|thumb|center|Structure and switching process of a NOT gate.<br>]] | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | |||
| - | + | ==Network construction== | |
| - | + | Designing complex biological networks based on either traditional protein engineering or our new bioLOGICS is still a complex task. As described above our bioLOGICS design allows the creation and precise connection of logic gates. To illustrate how a bioLOGICS network is put together we developed a software allowing the fast construction of a custom-made network.<br> | |
| - | + | To read more about this, take a look at our [https://2010.igem.org/Team:TU_Munich/Software software page] | |
| - | + | =Our Objective= | |
| - | + | Putting the implementation described above into practice, will be a major challenge. For this year's iGEM competition our goal is to do the first step: design and build a switch that can be toggled by a RNA molecule. To be precise, we want to apply the design rules of our switch to modify a transcription terminator in such a way that it interacts with a second RNA molecule and, as a result, is no longer capable of forming a stem loop. This objective will require intensive ''in silico'' designing and modeling of switches based on different terminators and their corresponding transmitters. In connection to this theoretical part, we also have to test and verify the switches. For this step, we establish custom-made assays, ''in vitro'' and ''in vivo''. | |
| - | + | ||
| - | = | + | |
| - | + | ||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | + | Once the objective mentioned above is accomplished, these basic RNA/RNA-interactions have to be modified in such a manner that the described identity/trigger site pattern for the transmitter and the complementary recognition/target site switch composition has to be established. The most important requirement is to is to optimize these modules that the transmitter is only able to switches specifically, meaning only in the presence of both, identity AND trigger site. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
Once the objective mentioned above is accomplished, the creation of an OR gate will be rather simple since it only requires two switches. However the creation of an AND or NOT gate and optimizing the logic gates to improve their responds function will remain the goal of future work. Also the creation of small networks and the correct integration of BioBricks as input and output molecules will be future challenges. Furthermore, we wanted to rather focus on the development and the testing of our structural design of the switches, rather than developing a variety of new BioBricks. | Once the objective mentioned above is accomplished, the creation of an OR gate will be rather simple since it only requires two switches. However the creation of an AND or NOT gate and optimizing the logic gates to improve their responds function will remain the goal of future work. Also the creation of small networks and the correct integration of BioBricks as input and output molecules will be future challenges. Furthermore, we wanted to rather focus on the development and the testing of our structural design of the switches, rather than developing a variety of new BioBricks. | ||
==''In silico'' design== | ==''In silico'' design== | ||
| - | We used [[Team:TU_Munich/Project#in silico design |''in silico'' | + | As described above, our switches are based on certain design rules. However, there still are different structural parameters that need to be tested and optimized (length of recognition site and target site, choice of terminator, etc.). |
| + | We used [[Team:TU_Munich/Project#in silico design |''in silico'' design]] and [[Team:TU_Munich/Modeling| modeling]]) to test different parameters. Furthermore we tried to use the [[Team:TU_Munich/Glossary#Antitermination|antitermination principle]] observed in nature, such as [[Team:TU_Munich/Glossary#Attenuation| attenuation]] in ''E. coli'' or [[Team:TU_Munich/Glossary#Tiny Abortive RNA´s| tiny abortive RNA´s]] of T7-phage. | ||
==Evaluation and Measurements== | ==Evaluation and Measurements== | ||
To evaluate the functionality of our molecular switches, we first had to establish several assays. Therefore, we improved an existing [[Team:TU_Munich/Lab#In vivo Measurements |''in vivo'' assay]] and developed an [[Team:TU_Munich/Lab#In vitro Transcription | ''in vitro'' assay]] for this purpose. For more information please refer to the [[Team:TU_Munich/Lab | lab]] section. | To evaluate the functionality of our molecular switches, we first had to establish several assays. Therefore, we improved an existing [[Team:TU_Munich/Lab#In vivo Measurements |''in vivo'' assay]] and developed an [[Team:TU_Munich/Lab#In vitro Transcription | ''in vitro'' assay]] for this purpose. For more information please refer to the [[Team:TU_Munich/Lab | lab]] section. | ||
| + | <br> | ||
| + | <br> | ||
| + | Summarizing, the main challenges are | ||
| + | * to find a suitable terminator construct and design a complementary trigger unit, which is only functional in combination with a specificity site - meaning an optimization of the '''thermodynamically parameters''' (see[[Team:TU_Munich/Project#in silico design| in silico design]]) | ||
| + | * to investigate whether the transmitter/switch interaction reaction is on a timescale to be competitive to terminator formation - meaning an comparison of '''kinetic parameters''' (see [[Team:TU_Munich/Modeling|Modeling page]]) | ||
| + | * to proof antitermination can be also be caused by synthetically RNA-interaction (see [[Team:TU_Munich/Glossary#Antitermination| Antitermination in nature]] and [[Team:TU_Munich/Project#Results| ''in vivo'' and ''in vitro'' measurements]] ) | ||
| + | |||
| + | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
=Results= | =Results= | ||
| Line 166: | Line 184: | ||
==in silico Design of Switching and Trigger Unit== | ==in silico Design of Switching and Trigger Unit== | ||
| - | + | As described on the [[Team:TU_Munich/Project | project]] page, one key aspect of our switches is the idea, that a [[Team:TU_Munich/Glossary#Transmitter_(bioLOGICS) | RNA transmitter molecule]] is capable to shift the state of a switch only if its [[Team:TU_Munich/Glossary#Trigger_Site_(bioLOGICS) | trigger site]] is present and its [[Team:TU_Munich/Glossary#Identity_Site_(bioLOGICS) | identity site]] corresponds to the [[Team:TU_Munich/Glossary#Recognition_Site_(bioLOGICS) | recognition site]] of the [[Team:TU_Munich/Glossary#Switch_(bioLOGICS) | switch]]. We successfully constructed several switches and their corresponding transmitter RNA ''in silico'' on a thermodynamical basis. We modified different transcriptional terminators in such a way, that the formation of the terminator was prevented by a transmitter molecule. As desired, this only occured if the transmitter molecule contained both, a trigger and an identity site. Analogously, we were able to design and verify a NOT gate using the same thermodynical approach. | |
| - | + | ||
| - | + | ||
| - | + | ==Diffusion and RNA Folding Dynamics== | |
| - | + | We estimated the diffusion time for our constructs and modeled the folding dynamics of our bioLOGICS switches including the switching process with a stochastic RNA folding program. We were able to provide better insight in their folding dynamics and proved that they are able to interrupt termination. We also optimized the switches and the corresponding signals. Furthermore, we combined the switches what resulted in a logic gate. See our [[Team:TU Munich/Modeling|Modeling page]] for further details. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | We | + | |
==''in vivo'' Functionality Screening== | ==''in vivo'' Functionality Screening== | ||
| - | Since our logic gates are intended to function in living cells, ''in vivo'' measurements were essential. In a set of experiments we concentrated on two different | + | Since our logic gates are intended to function in living cells, ''in vivo'' measurements were essential. In a set of experiments we concentrated on two different switches based on known [[Team:TU_Munich/Glossary#Attenuation|attenuators]] from nature: the [[Team:TU_Munich/Modeling#Switch|HisTerm]] and [[Team:TU_Munich/Modeling#Switch|TrpTerm]]. Focusing on fluorescent proteins for quantifiable input and output we designed a functional and robust screening system. For greater detail see [[Team:TU_Munich/Lab#Experiment_Design|Experimental Design]]. Unfortunately, setting up a working screening system failed twice. Only in redesigning and improving the screening plasmid pSB1A10 we succeeded, but lost precious time. |
| - | Ultimately, the two switches displayed remarkable differences in their terminator efficiency, but neither of them responded to their corresponding signal. However, screening one transmitter signal | + | Ultimately, the two switches displayed remarkable differences in their terminator efficiency, but neither of them responded to their corresponding signal. However, screening one transmitter signal does not disprove the basic working principle of our system. Limited by time, we hope for future teams to take up our work and to use our improved test system that we submitted to the parts registry, for performing successful in vivo measurement. |
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | Considering the high complexity of ''in vivo'' measurements compared to other experimental challenges, a robust and easy to handle test system for PoPS-based devices | + | Considering the high complexity of ''in vivo'' measurements compared to other experimental challenges, a robust and easy to handle test system for [[Team:TU_Munich/Glossary#PoPS-based devices| PoPS-based devices]] is desirable. As described in [[Team:TU_Munich/Lab#Experiment_Design|Experimental design]], we used fluorescent proteins: RFP or mCherry to measure the amount of produced output and eGFP for normalization. Our first attempt, using the screening plasmid pSB1A10, yielded no interpretable results. Switching the fluorescent protein to mCherry did not work either, but after several experimental setups we determined a transcriptional problem causing no reporter protein expression regardless of the inserted part. Thereby we demonstrated the screening plasmid pSB1A10 to be [[Team:TU_Munich/Biobricks#Falsification| malfunctioning]]. |
| - | Finally a new design based on pSB1A10 lead to a functional and robust screening system. A second promoter with identical induction properties inside the BioBrick cloning site enforces transcription of the PoPS-based device and the mCherry output. | + | Finally a new design based on pSB1A10 lead to a functional and robust screening system (compare [[Team:TU_Munich/Parts#Screening system: Backbone BBa_K494001| Screening system: Backbone BBa_K494001]]). A second promoter with identical induction properties inside the BioBrick cloning site enforces transcription of the PoPS-based device and the mCherry output. |
| - | Exemplary, the graph below on the right shows the positive control, induced and uninduced at OD<sub>600</sub>=0.7 followed by 16 h incubation at 25 °C. Clearly visible are eGFP and mCherry fluorescence in the induced samples. The uninduced control showed no fluorescence at all, demonstrating the PBad promoter to be tight and providing | + | Exemplary, the graph below on the right shows the positive control, induced and uninduced at OD<sub>600</sub>=0.7 followed by 16 h incubation at 25 °C. Clearly visible are eGFP and mCherry fluorescence in the induced samples. The uninduced control showed no fluorescence at all, demonstrating the PBad promoter to be tight and providing very low basal transcription, what is a major advantage for the screening system. This newly designed screening approach renders the characterization of PoPS-based devices in general and switches in particular easy and robust. The low basal transcription furthermore fulfills one of the most important requirements for the designed switches, since output transmitters may only be produced in presence of an input transmitter. This helps to avoid strong "background" noise, which would extremely harden the successful interconnection of several switches. |
<br> | <br> | ||
[[Image:TUM2010_PosControlklein.JPG|200px||thumb|left|Bacteria containing positive control]] | [[Image:TUM2010_PosControlklein.JPG|200px||thumb|left|Bacteria containing positive control]] | ||
[[Image:TUM2010_graphPosControl1.png|355px|thumb|center|Emission spectra of induced (green/red) and uninduced(black) positive control BBa_K494002 ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | [[Image:TUM2010_graphPosControl1.png|355px|thumb|center|Emission spectra of induced (green/red) and uninduced(black) positive control BBa_K494002 ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | ||
<br> | <br> | ||
| - | Due to the time limitations of the iGEM completion we had to focus our efforts on few | + | Due to the time limitations of the iGEM completion we had to focus our efforts on few switches after designing the screening system. Relying on the functionality of systems occurring in nature, we choose the [[Team:TU_Munich/Modeling#Switch|HisTerm]] as well as the [[Team:TU_Munich/Modeling#Switch|TrpTerm]]. Both switches are based on known natural [[https://2010.igem.org/Team:TU_Munich/Glossary#Attenuation|attenuators]]. Testing synthetic and none-naturally switchable terminators in vivo are goals for future work. |
| - | Delorme et al. reported the His-Terminator to be a remarkable effective Terminator with more than 99% termination | + | Delorme et al. reported the His-Terminator to be a remarkable effective Terminator with more than 99% termination efficiency.<sup>[[Team:TU_Munich/Project#ref12|[12]]]</sup> The exemplary measurement below on the right confirms the high terminator efficiency. In fact, we could not detect any mCherry fluorescence in any cells containing the [[Team:TU_Munich/Modeling#Switch|HisTerm]]. Even induction of the corresponding signal transmitter RNA via IPTG did not alter the Terminator efficiency. Again time was the limiting factor and prevented us from testing more than one corresponding transmitter, although the [[Team:TU_Munich/Modeling| Modeling]] highly suggested the necessarily of finding an optimized transmitter length. Thus, the results are insufficient either to prove or to disprove the functionality of the [[Team:TU_Munich/Modeling#Switch|HisTerm]] or our concept in general. |
<br> | <br> | ||
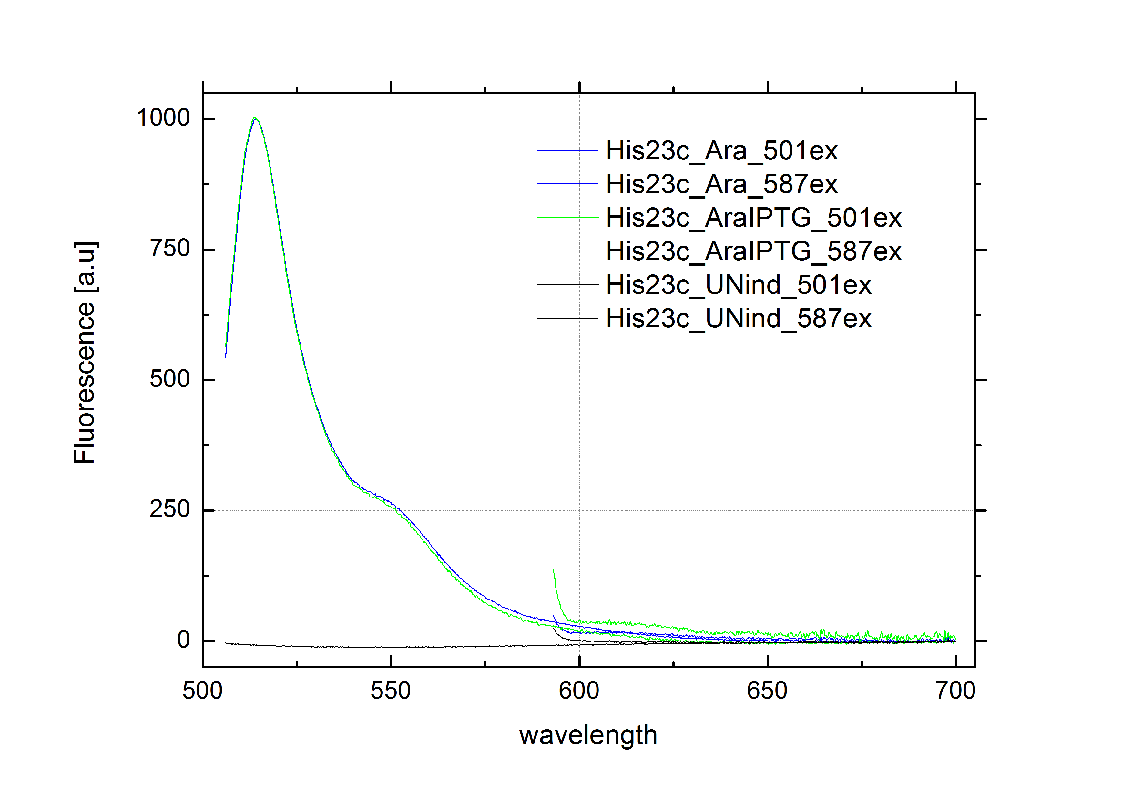
[[Image:TUM2010_HisSwitchklein.JPG|200px|thumb|left|Bacteria containing HisTerm]][[Image:TUM2010_HisSwitchGraph1.png|355px|thumb|center|Emission spectra of induced and uninduced screening plasmid BBa_K494002 containing HisTerm ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | [[Image:TUM2010_HisSwitchklein.JPG|200px|thumb|left|Bacteria containing HisTerm]][[Image:TUM2010_HisSwitchGraph1.png|355px|thumb|center|Emission spectra of induced and uninduced screening plasmid BBa_K494002 containing HisTerm ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | ||
<br> | <br> | ||
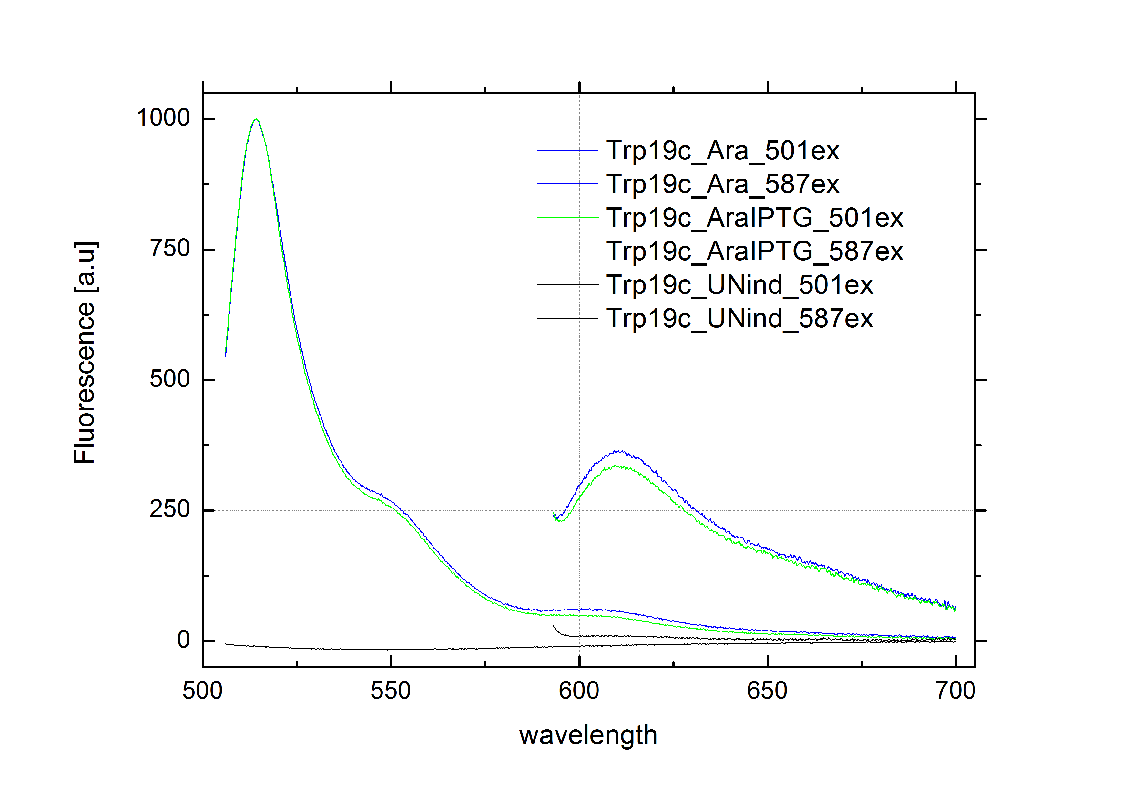
| - | Attaining only 90% terminator | + | Attaining only 90% terminator efficiency, the natural Trp [[https://2010.igem.org/Team:TU_Munich/Glossary#Attenuation|Attenuator]] is known be less effective than the [[Team:TU_Munich/Modeling#Switch|HisTerm]].<sup>[[Team:TU_Munich/Project#ref13|[13]]]</sup> The graph on the right depicts our designed [[Team:TU_Munich/Modeling#Switch|TrpTerm]] characteristic efficiency of about 40 %, notably below the natural standard. Allowing 60% transcription in the “off” state excludes the [[Team:TU_Munich/Modeling#Switch|TrpTerm]] from possible candidates for a scalable network of logic gates, due to the mentioned required "yes or no" function (see [[Team:TU_Munich/Project#Implementation| Implementation and how to connect Biobricks]]). Thus the [[Team:TU_Munich/Modeling#Switch|TrpTerm]] is inoperative as intended, but may still be useful in other contexts. Similar to the [[Team:TU_Munich/Modeling#Switch|HisTerm]], the [[Team:TU_Munich/Modeling#Switch|TrpTerm]] also did not react to the induction of the corresponding signal. Under circumstances, termination efficiencies altered by the transmitter are on a low range and not resolvable within observed 40% basal transcription. |
<br> | <br> | ||
[[Image:TUM2010_TrpSwitchklein.JPG|200px|thumb|left|Bacteria containing TrpTerm]][[Image:TUM2010_TrpSwitchGraph1.png|355px|thumb|center|Emission spectra of induced and uninduced screening plasmid BBa_K494002 containing TrpTerm ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | [[Image:TUM2010_TrpSwitchklein.JPG|200px|thumb|left|Bacteria containing TrpTerm]][[Image:TUM2010_TrpSwitchGraph1.png|355px|thumb|center|Emission spectra of induced and uninduced screening plasmid BBa_K494002 containing TrpTerm ; green: eGFP fluorescence ex: 501 nm, red: mCherry fluorescence ex: 587 nm]] | ||
<br> | <br> | ||
| - | Making use of our improved screening system we also carried out some ''in vivo'' kinetic measurements in addition to the end-point measurements above. In contrast to the ''in vitro'' experiments we did not obtain significant results for the | + | Making use of our improved screening system we also carried out some ''in vivo'' kinetic measurements in addition to the end-point measurements above. In contrast to the ''in vitro'' experiments we did not obtain significant results for the characterization of our switches. As the switching process is many times faster than protein synthesis our ''in vivo'' kinetics include the synthesis of mCherry as well as its maturation. Therefore we centered our attention on end-point experiments. For more information browse the [[Team:TU_Munich/Lab#Lab_Book|lab book]]. <br> |
Considering our ''in vivo'' measurements, neither of the tested switches showed any effect regarding the signal induction. But due to the small number of tested switches and signals this can hardly be regarded as disprove of concept. In particular in light of the recent findings by Sooncheol proving antitermination in principle using a T7 system.<sup>[[Team:TU_Munich/Project#ref14|[14]]]</sup> | Considering our ''in vivo'' measurements, neither of the tested switches showed any effect regarding the signal induction. But due to the small number of tested switches and signals this can hardly be regarded as disprove of concept. In particular in light of the recent findings by Sooncheol proving antitermination in principle using a T7 system.<sup>[[Team:TU_Munich/Project#ref14|[14]]]</sup> | ||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| - | ==in vitro Screening== | + | ==''in vitro'' Screening== |
| + | To minimize the amount of disturbing factors we decided to countercheck our ''in vivo'' results with a set of ''in vitro'' measurements. While the ''in vitro'' systems are no doubt much less complex than living cells, the work with these set-ups proved to be quite as difficult. | ||
| + | Just as with the ''in vivo'' measurements we could prove our switching system neither right nor wrong, leaving enough work for future iGEM teams. | ||
| + | |||
{{:Team:TU Munich/Templates/ToggleBoxStart}} | {{:Team:TU Munich/Templates/ToggleBoxStart}} | ||
| - | ===in vitro translation=== | + | ===''in vitro translation''=== |
| - | + | ||
| - | Beside optimization of the reporter proteins in use, | + | Beside optimization of the reporter proteins in use, the major problem occuring in the experiments was the low capacity of the kit. The signal intensity was very low, which made it difficult to observe any signal intensity alterations, so no conclusion could be drawn from these measurements. |
| + | |||
| + | ===''in vitro'' transcription=== | ||
| + | We used two completely independent ''in vitro'' systems: Using ''E.coli'' RNA Polymerase we analyzed the His and Trp switches that had already been tested ''in vivo''. In a second set-up, we used the well-established T7 RNA Polymerase and switch based on the T7 terminator as well as several signal sequences. | ||
| + | |||
| + | ====T7 System==== | ||
| + | In contradiction to the results of Kang and coworkers and other groups, in our ''in vitro'' set-up the T7 terminator did not seem to terminate at all. The negative control (Promoter_Terminator_malachite binding aptamer) showed a similar increase in fluorescence as the positive control (Promoter_random sequence_malachite binding aptamer). | ||
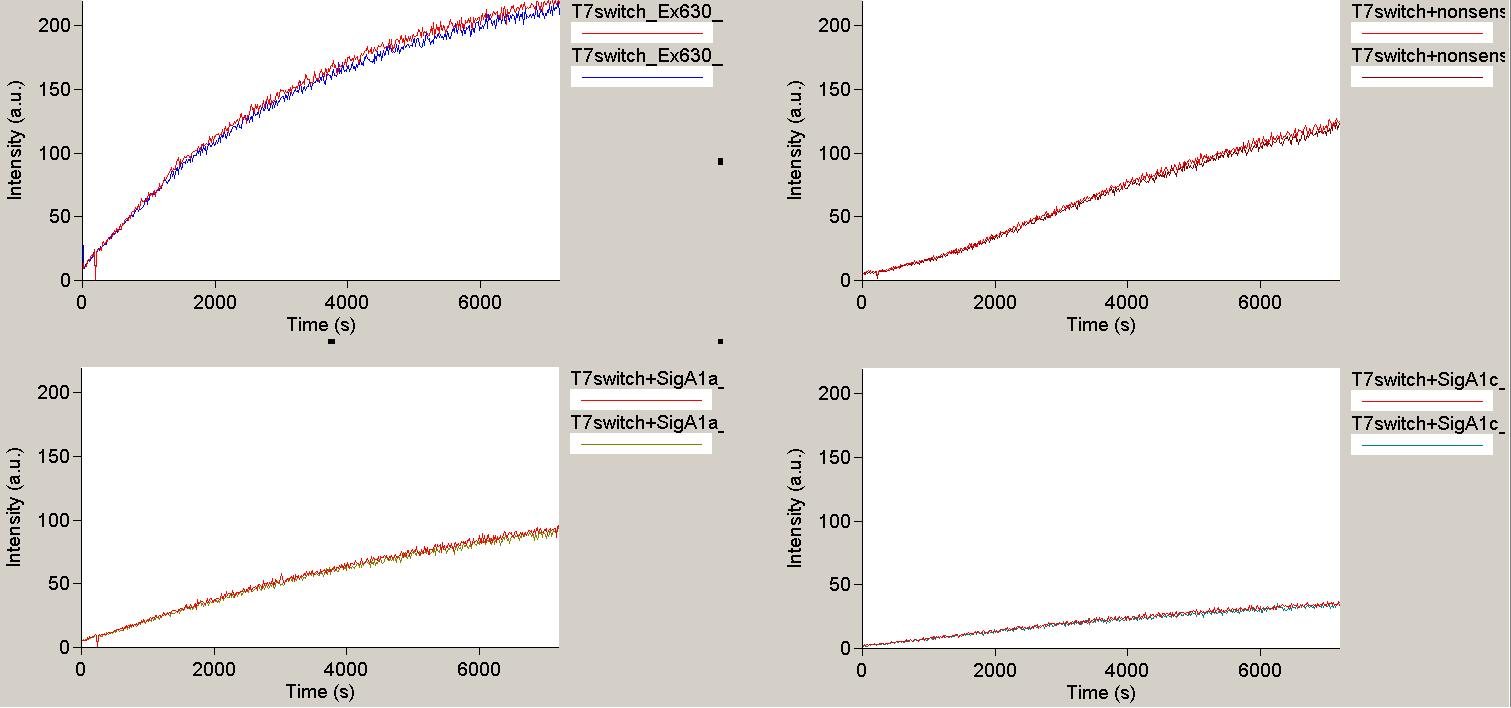
| + | [[Image:TUM2010_T7Result1.png|350px||thumb|left|''in vitro'' transcription measurement of T7 terminator with no signal(upper left), nonsense signal (upper right) and two different designed signals (below)]] | ||
| + | [[Image:TUM2010_T7Result3.png|350px||thumb|right|''in vitro'' transcription measurement of positive control(upper left and T7 terminator with three different designed signals (remaining traces)]]<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Furthermore denaturing Polyacrylamide Gel Electrophoresis (PAGE) confirmed that there was no observeable termination of transcription. The addition of a signaling sequence led to a significantly lower increase in fluorescence, which can be attributed to the fact that both DNA sequences, switch and signal, compete for RNA Polymerases. | ||
| + | However, there is almost no difference between the designed signals and random sequences, which is not a big surprise since there can be no antitermination if the terminator itself does not work.<br> | ||
| + | |||
| + | Possible explanations for the contradiction between our results and those of Kang and coworkers might be the experimental set-up and the RNA Polymerases we used. Different variants of T7 RNA Polymerase might respond in different ways to terminator structures, and the termination might be influenced by the presence or absence of cofactors, depending on the purification methods used in producing the Polymerase.<br><br> | ||
| + | |||
| + | This set-up offers a lot of possible experiments for the future, which we would have loved to conduct with a just a bit more time... | ||
| + | |||
| + | ====''E.coli'' System==== | ||
| + | |||
| + | Compared to the T7 System, the ''E. coli'' RPO system produced poor increases in fluorescence, indicating little RNA synthesis. It was shown that the presence of a terminator decreases, as expected, the production of downstream RNA.<br> | ||
| + | <br> | ||
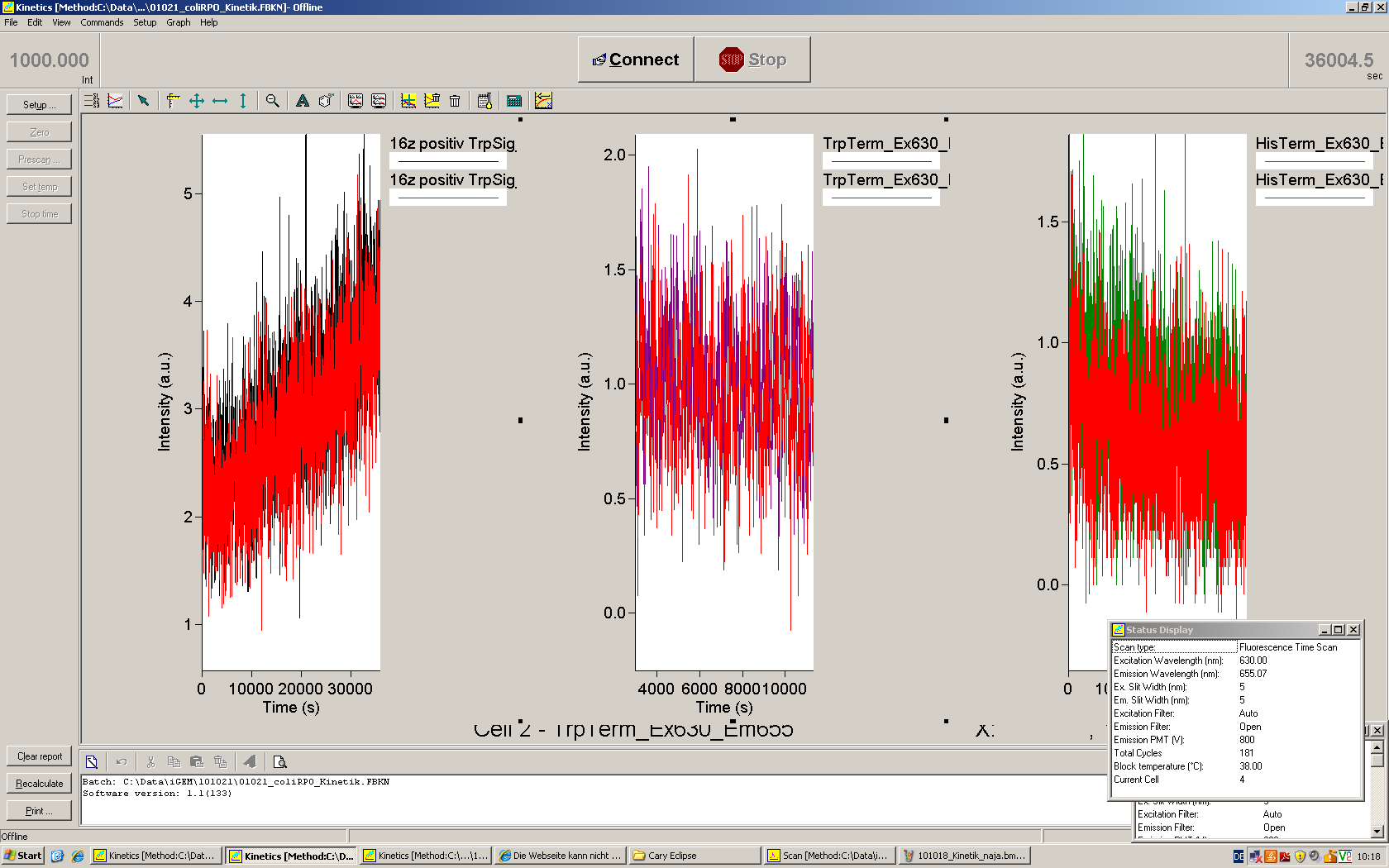
| + | [[Image:TUM2010_101023kinetik.PNG|350px||thumb|left|''in vitro'' transcription measurement of Switch TrpTerm (upper traces) and positive control (lower traces). Left side: with Trp-signal, right side: no signal]] | ||
| + | [[Image:TUM2010_101022_Kinetik.png|350px||thumb|right|''in vitro'' transcription measurement of positive control (left), Switch TrpTerm (center) and switch HisTerm(right) | ||
| + | <br> | ||
| + | ]]<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | This result was also confirmed by denaturing PAGE. However, due to the poor changes in fluorescence we were not able to actually characterize the behaviour of our switches ''in vitro'', and the small RNA concentrations did not allow a quantitative interpretation of our gels. | ||
| + | |||
| + | <br> | ||
| + | |||
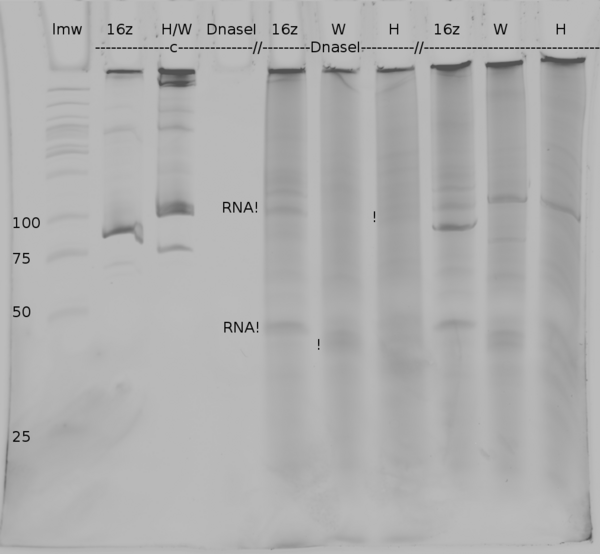
| + | [[Image:TUM2010_coliGel.png|500px||thumb|center|denaturing polyacrylamide gel electrophoresis of DNaseI digested samples from ''in vitro'' transcription of positive control (16z), Switch TrpTerm (W) and switch HisTerm(H). c marks the lanes in which the DNA was injected, the last three lanes show the undigested samples]] | ||
| + | |||
| + | A major problem with this method was the low concentration of the ordered Polymerase resulting in a much weaker overall signal as comparable measurements using the T7 Polymerase. <br><br> | ||
| + | In future experiments we might try to work with smaller volumes in order to reach higher concentration of RPO and of the synthesized RNA molecules, so measuring in 96 well plate readers might be a good choice. | ||
| + | |||
| - | |||
| - | |||
| - | |||
{{:Team:TU Munich/Templates/ToggleBoxEnd}} | {{:Team:TU Munich/Templates/ToggleBoxEnd}} | ||
| Line 307: | Line 271: | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=References= | =References= | ||
| Line 317: | Line 278: | ||
<html><a name="ref3"></a></html>[3] https://2009.igem.org/Team:TUDelft | <html><a name="ref3"></a></html>[3] https://2009.igem.org/Team:TUDelft | ||
<html><a name="ref4"></a></html>[4] https://2008.igem.org/Team:Heidelberg | <html><a name="ref4"></a></html>[4] https://2008.igem.org/Team:Heidelberg | ||
| - | <html><a name="ref5"></a></html>[5] | + | <html><a name="ref5"></a></html>[5] Maung Nyan Win and Christina D. Smolke, Science Oct. 2008 Vol. 322. no. 5900, pp. 456 - 460 |
| - | <html><a name="ref6"></a></html>[6] | + | <html><a name="ref6"></a></html>[6] Lu, T.K., A.S. Khalil, and J.J. Collins, Next-generation synthetic gene networks. Nature biotechnology, 2009. 27(12): p. 1139-1150. |
| - | <html><a name=" | + | <html><a name="ref7"></a></html>[7] Schaller, R.R., Moore's law: past, present and future. Spectrum, IEEE, 2002. 34(6): p. 52-59. |
| - | <html><a name=" | + | <html><a name="ref8"></a></html>[8] von Mering, C., et al., Comparative assessment of large-scale data sets of protein–protein interactions. Nature, 2002. 417(6887): p. 399-403. |
| - | <html><a name=" | + | <html><a name="ref9"></a></html>[9] Mandal, M. and R.R. Breaker, Gene regulation by riboswitches. Nature Reviews Molecular Cell Biology, 2004. 5(6): p. 451-463. |
| - | <html><a name=" | + | <html><a name="ref10"></a></html>[10] Benner, S.A. and A.M. Sismour, Synthetic biology. Nature Reviews Genetics, 2005. 6(7): p. 533-543. |
| - | <html><a name=" | + | <html><a name="ref11"></a></html>[11] Beaudry, A. and G. Joyce, Directed evolution of an RNA enzyme. Science, 1992. 257(5070): p. 635-641. |
<html><a name="ref12"></a></html>[12] Delorme, Ehrlich and Renault, Regulation of Expression of the Lactococcus lactis Histidine Operon. Journal of Bacteriology, Apr. 1999, p. 2026–2037 | <html><a name="ref12"></a></html>[12] Delorme, Ehrlich and Renault, Regulation of Expression of the Lactococcus lactis Histidine Operon. Journal of Bacteriology, Apr. 1999, p. 2026–2037 | ||
<html><a name="ref13"></a></html>[13] Trun and Trempy(2003): Fundamental Bacterial Genetics, Wiley-Blackwell, Chapter 12 | <html><a name="ref13"></a></html>[13] Trun and Trempy(2003): Fundamental Bacterial Genetics, Wiley-Blackwell, Chapter 12 | ||
<html><a name="ref14"></a></html>[14]Sooncheol Lee, Huong Minh Nguyen and Changwon Kang, Tiny abortive initiation transcripts exert antitermination activity on an RNA hairpin-dependent intrinsic terminator. Nucleic Acids Research, 2010, 1–9 | <html><a name="ref14"></a></html>[14]Sooncheol Lee, Huong Minh Nguyen and Changwon Kang, Tiny abortive initiation transcripts exert antitermination activity on an RNA hairpin-dependent intrinsic terminator. Nucleic Acids Research, 2010, 1–9 | ||
| - | |||
| - | |||
<!-- The idea behind our project is to change the way BioBricks have been used up to now. Over the years, many receptors and signals have been constructed as BioBricks during the annual iGEM competition, but still it is not possible to interconnect these Bricks in a complex biological network resuting in a cell, that is able to respond to its environment giving differenciated responses depending on the input signals. (Beispiel: cambridge hat das gemacht, xx dies, aber eine zelle kann nicht beides...<br> | <!-- The idea behind our project is to change the way BioBricks have been used up to now. Over the years, many receptors and signals have been constructed as BioBricks during the annual iGEM competition, but still it is not possible to interconnect these Bricks in a complex biological network resuting in a cell, that is able to respond to its environment giving differenciated responses depending on the input signals. (Beispiel: cambridge hat das gemacht, xx dies, aber eine zelle kann nicht beides...<br> | ||
Latest revision as of 03:49, 28 October 2010
|
||||||||
|
|
VisionUntil today, 13.628 biobrick sequences[1] have been submitted to partsregistry, thereof 102 reporter units and 12 signaling bricks.
Since then, people are trying to arrange these single biological building blocks in such a manner that allows producing special biotechnological products (metabolic engineering), developing biological sensory circuits (biosensors) and even giving microorganisms the ability to react on multiple environmental factors and serve both as disease indicator and drug. These examples and further promising ideas were implemented on previous iGEM-competitions.[2][3][4] Generally speaking, the above adapter has to meet the following requirements:
ImplementationTo functionally connect BioBricks, there are several possibilities including genetic switches, riboswitches and direct protein-protein interactions. We investigated several hypothetically principles, and decided to focus our practical work on the development of a RNA-RNA interaction-based switch. These switches are capable of changing between two states, a state of antitermination and termination, and make use of highly-specific RNA-RNA interaction. In principle such a switch can fulfill all requirements mentioned previously. The following text clarifies how these switches work in detail. How to connect BioBricksOur adapter is a system, that activates or disables BioBricks (output BioBricks) in response to the presence of other Biobricks (input Biobricks). Our approach uses a molecular network to put this into practice and consists of four major elements:
In order to connect different BioBricks, our network requires four major types of components:
Computer vs. molecular network - and our approach
Logic gates in a molecular network are often compared to transistors used in a computer, where billions of transistors are incorporated[7]. The main advantage on a computer chip is, all transistors share the same functional principle, and only the way connecting them in a special sequence allows specific addressing of only a subset of other transistors by an input. However, spatially fixed connections of molecular logic gates are not possible in a living cell. The "wiring" within a cell relies on the specific interaction between transmitter molecule and their corresponding logic gates, for example implemented by protein-protein/ligand-protein interactions or specific ligand-riboswitch interactions.[8][9] As a result, in a cell, each occurring logic gate ("transistor") has to be different, at least in a special recognition site[10] - for example like different transcription factors, recognizing different DNA-sites. Thanks to evolution, nature easily can invent a new transistor for each task - science achieves this only on a limited scale, and producing synthetic molecular logic gates artificially by either rational or evolutionary protein or riboswitch engineering, is limited to small circuits so far[11]. Our project aims to establish a molecular switch as close as possible to a electronic transistor, thus sharing the same functional principle for all logic gates. At the same time, we want to design a easily exchangeable recognition site, which can individually be designed by everyone! These elements can be combined to build up a molecular network (see illustration). Each input molecule (such as a BioBrick) produces a unique transmitter molecule. All transmitters belong to the same type of molecule and share a common design. However, each transmitter molecule can only interact and activate a certain subset of logic gates. In other words, logic gates have to recognize as well as bind the corresponding transmitter molecules and are capable of producing a new output transmitter molecule. Depending on the type of the logic gate (AND, OR or NOT[6]), an output transmitter is only created if both input transmitter molecules are present (AND), at least one of two input transmitters is present (OR) or if no input transmitter is present at all (NOT). Once a logic gate has produced a new output transmitter, these transmitters can in turn address another subset ("layer") of logic gates. In theory many layers of logic gates can be connected this way allowing the creation of large networks. Until this step, various transmitter molecules might have been produced. But in order to create a Biobrick output, the last layer of logic gates finally generates transmitter molecules that will not active logic gates, but will rather interact with the cell metabolism to produce a BioBrick response. In other words, the last layer of transmitter molecules is capable of regulating BioBrick formation.
Design and functional principle of logic gatesThe concept introduced above provides a framework that can potentially serve as an universal adapter between different BioBricks. However, the logic gates have not been specified more precisely so far. This will be done in the following section.
Generally speaking, our logic gates are to possess the following characteristics:
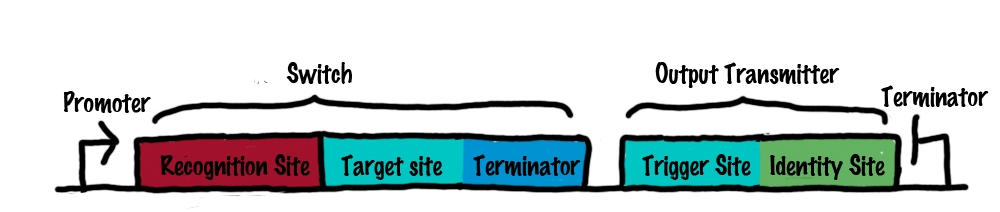
In order to build logic gates for our bioLOGICS system we will first create a simple switch. A switch can be activated by one transmitter RNA and produce an output transmitter RNA. In contrast to a logic gate, a switch does not perform logic operations. However by combining switches, logic gates can be created. The following text will first describe how the developed switch works and secondly, how logic gates such as AND/OR/NOT can be created using these switches.  A The basic structure of a bioLOGICS switch (left) and a transmitter molecule (right). BThe process of switching. See the text in the close-by "Read more" section for details. Rectangles present the composition of our functional units on the level of DNA. Fringed lines represent RNA produced by RNA polymerase. The stem loop structure depicts the switchable terminator. Terminator and target site are illustrated in blue and turquoise, respectively. Recognition sites are indicated in different colors, in this case red for the input transmitter and green for the output transmitter.Each switch and or later logical unit has to be flanked by a promotor and another constitutive terminator, to allow RNA-production by RNA-polymerase in a proper way.
SwitchRoughly speaking, a switch can be regarded as an enhanced switchable transcriptional terminator. The enhancement can be described easier by dividing a switch into its functional components:
Summing up, the recognition site allows a specific interaction between switches and transmitter molecules. Once this interaction is formed, an interaction between the transmitter and the target will actually switch the state of the terminator. This allows the specific arrangement and interconnection of numerous of these switches by transmitter molecules, without changing the target site. Comparable to wires connecting many identical transistors, our target site remains the same.
Transmitter RNA´sAs desccribed above, transmitter RNAs are the input and output of bioLOGICS switches (compare How to connect BioBricks). These transmitters are short ssRNA molecules representing the "trigger" to shift switches between the "on" and "off" state. To fulfill this role, they need to posses the following properties:
Again, these two properties are fulfilled by two components of the transmitter:
Summing up, we applied the principle introduced for the switches to the transmitter molecules. In contrast to previous approaches on this field [12], we introduced the described synthetic trigger site in such a manner that it is not able to change the state of the terminator on its own, but only in combination with the identity site. So the challenge is to arrange and optimize these elementary building blocks thermodynamically, that a trigger site is only able to switch in combination with its respective identity site. This was done by in silico design using NUPACK, presented in section in silico design.
Putting it all together: the switching processThe functional principle of the designed switches is illustrated in the figure. The switch is positioned on DNA upstream of a desired output transmitter. So in the absence of a triggering transmitter molecule, transcription will be canceled by the formation of a RNA stem loop in the nascent RNA-chain. This will cause the RNA polymerase to stop transcription and fall off the DNA and consequently no output RNA will be produced. This process only relies on rho-independent termination.On the other hand, in the presence of a input transmitter, this small functional RNA inhibits the stem loop formation by complementary base-pairing and hence avoids termination of transcription. In detail, the identity site (red part on transmitter) binds the recognition site (red part on switch) and serves as toehold, which will thermodynamically allow the trigger site (turquoise part on transmitter) to perform a strand displacement and open up the stem loop structure. Consequently the polymerase can read all the way through and form the output RNA. From switches towards bioLOGICS logic gatesAs described, each switch can be accessed by a specific RNA-transmitter molecule, representing the input. In turn, another RNA-transmitter molecule will be produced if the switch shifts its state. This output transmitter of one switch can serve as input transmitter for the next switch by meaningful selection and design of the respective recognition sites. This easily allows arranging several switches in specific sequences and faulty wiring - the corner stone of a logical network.
Network constructionDesigning complex biological networks based on either traditional protein engineering or our new bioLOGICS is still a complex task. As described above our bioLOGICS design allows the creation and precise connection of logic gates. To illustrate how a bioLOGICS network is put together we developed a software allowing the fast construction of a custom-made network. Our ObjectivePutting the implementation described above into practice, will be a major challenge. For this year's iGEM competition our goal is to do the first step: design and build a switch that can be toggled by a RNA molecule. To be precise, we want to apply the design rules of our switch to modify a transcription terminator in such a way that it interacts with a second RNA molecule and, as a result, is no longer capable of forming a stem loop. This objective will require intensive in silico designing and modeling of switches based on different terminators and their corresponding transmitters. In connection to this theoretical part, we also have to test and verify the switches. For this step, we establish custom-made assays, in vitro and in vivo.
Once the objective mentioned above is accomplished, these basic RNA/RNA-interactions have to be modified in such a manner that the described identity/trigger site pattern for the transmitter and the complementary recognition/target site switch composition has to be established. The most important requirement is to is to optimize these modules that the transmitter is only able to switches specifically, meaning only in the presence of both, identity AND trigger site.
In silico designAs described above, our switches are based on certain design rules. However, there still are different structural parameters that need to be tested and optimized (length of recognition site and target site, choice of terminator, etc.). We used in silico design and modeling) to test different parameters. Furthermore we tried to use the antitermination principle observed in nature, such as attenuation in E. coli or tiny abortive RNA´s of T7-phage. Evaluation and MeasurementsTo evaluate the functionality of our molecular switches, we first had to establish several assays. Therefore, we improved an existing in vivo assay and developed an in vitro assay for this purpose. For more information please refer to the lab section.
ResultsEvery network starts with a basic unit. While our declared aim is to enable networks allowing fine-tuning of gene expression beyond the regular on/off, exploring such an on/off switch/signal pair is the first step towards a functional network. We constructed several units and tested their efficiency, robustness and reproducibility in vivo, in vitro and in silico. Furthermore we developed a software which allows easy constructions of networks based on our designed logic gates. Conclusive elaboration of a few first RNA-based logic units is the major contribution of our iGEM team. in silico Design of Switching and Trigger UnitAs described on the project page, one key aspect of our switches is the idea, that a RNA transmitter molecule is capable to shift the state of a switch only if its trigger site is present and its identity site corresponds to the recognition site of the switch. We successfully constructed several switches and their corresponding transmitter RNA in silico on a thermodynamical basis. We modified different transcriptional terminators in such a way, that the formation of the terminator was prevented by a transmitter molecule. As desired, this only occured if the transmitter molecule contained both, a trigger and an identity site. Analogously, we were able to design and verify a NOT gate using the same thermodynical approach. Diffusion and RNA Folding DynamicsWe estimated the diffusion time for our constructs and modeled the folding dynamics of our bioLOGICS switches including the switching process with a stochastic RNA folding program. We were able to provide better insight in their folding dynamics and proved that they are able to interrupt termination. We also optimized the switches and the corresponding signals. Furthermore, we combined the switches what resulted in a logic gate. See our Modeling page for further details. in vivo Functionality ScreeningSince our logic gates are intended to function in living cells, in vivo measurements were essential. In a set of experiments we concentrated on two different switches based on known attenuators from nature: the HisTerm and TrpTerm. Focusing on fluorescent proteins for quantifiable input and output we designed a functional and robust screening system. For greater detail see Experimental Design. Unfortunately, setting up a working screening system failed twice. Only in redesigning and improving the screening plasmid pSB1A10 we succeeded, but lost precious time. Ultimately, the two switches displayed remarkable differences in their terminator efficiency, but neither of them responded to their corresponding signal. However, screening one transmitter signal does not disprove the basic working principle of our system. Limited by time, we hope for future teams to take up our work and to use our improved test system that we submitted to the parts registry, for performing successful in vivo measurement. Considering the high complexity of in vivo measurements compared to other experimental challenges, a robust and easy to handle test system for PoPS-based devices is desirable. As described in Experimental design, we used fluorescent proteins: RFP or mCherry to measure the amount of produced output and eGFP for normalization. Our first attempt, using the screening plasmid pSB1A10, yielded no interpretable results. Switching the fluorescent protein to mCherry did not work either, but after several experimental setups we determined a transcriptional problem causing no reporter protein expression regardless of the inserted part. Thereby we demonstrated the screening plasmid pSB1A10 to be malfunctioning. Finally a new design based on pSB1A10 lead to a functional and robust screening system (compare Screening system: Backbone BBa_K494001). A second promoter with identical induction properties inside the BioBrick cloning site enforces transcription of the PoPS-based device and the mCherry output. Exemplary, the graph below on the right shows the positive control, induced and uninduced at OD600=0.7 followed by 16 h incubation at 25 °C. Clearly visible are eGFP and mCherry fluorescence in the induced samples. The uninduced control showed no fluorescence at all, demonstrating the PBad promoter to be tight and providing very low basal transcription, what is a major advantage for the screening system. This newly designed screening approach renders the characterization of PoPS-based devices in general and switches in particular easy and robust. The low basal transcription furthermore fulfills one of the most important requirements for the designed switches, since output transmitters may only be produced in presence of an input transmitter. This helps to avoid strong "background" noise, which would extremely harden the successful interconnection of several switches.
Attaining only 90% terminator efficiency, the natural Trp [[2]] is known be less effective than the HisTerm.[13] The graph on the right depicts our designed TrpTerm characteristic efficiency of about 40 %, notably below the natural standard. Allowing 60% transcription in the “off” state excludes the TrpTerm from possible candidates for a scalable network of logic gates, due to the mentioned required "yes or no" function (see Implementation and how to connect Biobricks). Thus the TrpTerm is inoperative as intended, but may still be useful in other contexts. Similar to the HisTerm, the TrpTerm also did not react to the induction of the corresponding signal. Under circumstances, termination efficiencies altered by the transmitter are on a low range and not resolvable within observed 40% basal transcription.
Making use of our improved screening system we also carried out some in vivo kinetic measurements in addition to the end-point measurements above. In contrast to the in vitro experiments we did not obtain significant results for the characterization of our switches. As the switching process is many times faster than protein synthesis our in vivo kinetics include the synthesis of mCherry as well as its maturation. Therefore we centered our attention on end-point experiments. For more information browse the lab book. in vitro ScreeningTo minimize the amount of disturbing factors we decided to countercheck our in vivo results with a set of in vitro measurements. While the in vitro systems are no doubt much less complex than living cells, the work with these set-ups proved to be quite as difficult. Just as with the in vivo measurements we could prove our switching system neither right nor wrong, leaving enough work for future iGEM teams.
in vitro translationBeside optimization of the reporter proteins in use, the major problem occuring in the experiments was the low capacity of the kit. The signal intensity was very low, which made it difficult to observe any signal intensity alterations, so no conclusion could be drawn from these measurements. in vitro transcriptionWe used two completely independent in vitro systems: Using E.coli RNA Polymerase we analyzed the His and Trp switches that had already been tested in vivo. In a second set-up, we used the well-established T7 RNA Polymerase and switch based on the T7 terminator as well as several signal sequences. T7 SystemIn contradiction to the results of Kang and coworkers and other groups, in our in vitro set-up the T7 terminator did not seem to terminate at all. The negative control (Promoter_Terminator_malachite binding aptamer) showed a similar increase in fluorescence as the positive control (Promoter_random sequence_malachite binding aptamer).
Furthermore denaturing Polyacrylamide Gel Electrophoresis (PAGE) confirmed that there was no observeable termination of transcription. The addition of a signaling sequence led to a significantly lower increase in fluorescence, which can be attributed to the fact that both DNA sequences, switch and signal, compete for RNA Polymerases.
However, there is almost no difference between the designed signals and random sequences, which is not a big surprise since there can be no antitermination if the terminator itself does not work. Possible explanations for the contradiction between our results and those of Kang and coworkers might be the experimental set-up and the RNA Polymerases we used. Different variants of T7 RNA Polymerase might respond in different ways to terminator structures, and the termination might be influenced by the presence or absence of cofactors, depending on the purification methods used in producing the Polymerase. This set-up offers a lot of possible experiments for the future, which we would have loved to conduct with a just a bit more time... E.coli SystemCompared to the T7 System, the E. coli RPO system produced poor increases in fluorescence, indicating little RNA synthesis. It was shown that the presence of a terminator decreases, as expected, the production of downstream RNA.
This result was also confirmed by denaturing PAGE. However, due to the poor changes in fluorescence we were not able to actually characterize the behaviour of our switches in vitro, and the small RNA concentrations did not allow a quantitative interpretation of our gels.
A major problem with this method was the low concentration of the ordered Polymerase resulting in a much weaker overall signal as comparable measurements using the T7 Polymerase.
SoftwareAlthough we could not show the full functionality of bioLOGICS in the lab we still want to demonstrate the potential of our approach. Hence we implemented the idea behind our logic gates in a program which illustrates how bioLOGCIS theoretically would allow the construction of complex information processing networks interconnecting BioBricks. For further details take a look at our Software page.
References[1] http://partsregistry.org/cgi/partsdb/Statistics.cgi [2] https://2009.igem.org/Team:Imperial_College_London/M1 encapsulation [3] https://2009.igem.org/Team:TUDelft [4] https://2008.igem.org/Team:Heidelberg [5] Maung Nyan Win and Christina D. Smolke, Science Oct. 2008 Vol. 322. no. 5900, pp. 456 - 460 [6] Lu, T.K., A.S. Khalil, and J.J. Collins, Next-generation synthetic gene networks. Nature biotechnology, 2009. 27(12): p. 1139-1150. [7] Schaller, R.R., Moore's law: past, present and future. Spectrum, IEEE, 2002. 34(6): p. 52-59. [8] von Mering, C., et al., Comparative assessment of large-scale data sets of protein–protein interactions. Nature, 2002. 417(6887): p. 399-403. [9] Mandal, M. and R.R. Breaker, Gene regulation by riboswitches. Nature Reviews Molecular Cell Biology, 2004. 5(6): p. 451-463. [10] Benner, S.A. and A.M. Sismour, Synthetic biology. Nature Reviews Genetics, 2005. 6(7): p. 533-543. [11] Beaudry, A. and G. Joyce, Directed evolution of an RNA enzyme. Science, 1992. 257(5070): p. 635-641. [12] Delorme, Ehrlich and Renault, Regulation of Expression of the Lactococcus lactis Histidine Operon. Journal of Bacteriology, Apr. 1999, p. 2026–2037 [13] Trun and Trempy(2003): Fundamental Bacterial Genetics, Wiley-Blackwell, Chapter 12 [14]Sooncheol Lee, Huong Minh Nguyen and Changwon Kang, Tiny abortive initiation transcripts exert antitermination activity on an RNA hairpin-dependent intrinsic terminator. Nucleic Acids Research, 2010, 1–9
|
|||||||
 "
"