Team:Imperial College London/Modelling/Protein Display/Objectives
From 2010.igem.org
(Difference between revisions)
m (proof read) |
m |
||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
| - | {{:Team:Imperial_College_London/Templates/ | + | {{:Team:Imperial_College_London/Templates/ModellingHeaderD}} |
{{:Team:Imperial_College_London/Templates/ModellingDetectionHeader}} | {{:Team:Imperial_College_London/Templates/ModellingDetectionHeader}} | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| Line 7: | Line 7: | ||
| | | | ||
<html> | <html> | ||
| - | The motivation to develop this model came from our design process. The idea of having surface proteins that could be cleaved and would then activate the receptor was very innovatory. However, with this new approach few questions arose that could not be answered without using computer models. Hence, the following aims for this model were specified: | + | The motivation to develop this model came from our design process. The idea of having surface proteins that could be cleaved and would then activate the receptor was very innovatory. However, with this new approach a few questions arose that could not be answered without using computer models. Hence, the following aims for this model were specified: |
<ol> | <ol> | ||
<li>Determine the concentration of Schistosoma elastase or TEV protease that should be added to the bacteria in order to trigger a response.</li> | <li>Determine the concentration of Schistosoma elastase or TEV protease that should be added to the bacteria in order to trigger a response.</li> | ||
| Line 14: | Line 14: | ||
</ol> | </ol> | ||
</html> | </html> | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:404px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:IC_Overview_Detection.jpg|400px]] | ||
| + | |- | ||
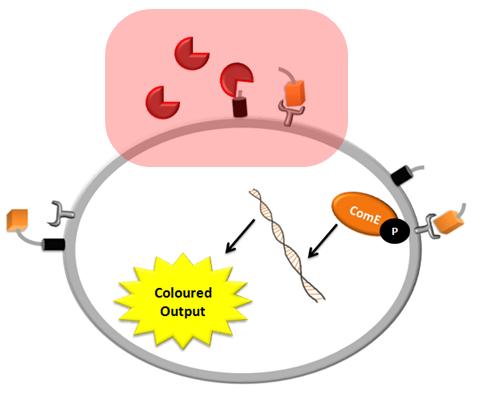
| + | |The pink shadow indicates which part of the whole system the detection model is simulating. It includes TEV or Schistosoma protease cleaving the surface protein to activate the receptor. | ||
| + | |} | ||
| + | </div> | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Protein_Display/Detailed_Description | Click here for a detailed description of this model...]] | ||
|} | |} | ||
Latest revision as of 03:33, 28 October 2010
| Modelling | Overview | Detection Model | Signaling Model | Fast Response Model | Interactions |
| A major part of the project consisted of modelling each module. This enabled us to decide which ideas we should implement. Look at the Fast Response page for a great example of how modelling has made a major impact on our design! | |
| Objectives | Description | Results | Constants | MATLAB Code |
| Objectives |
|
The motivation to develop this model came from our design process. The idea of having surface proteins that could be cleaved and would then activate the receptor was very innovatory. However, with this new approach a few questions arose that could not be answered without using computer models. Hence, the following aims for this model were specified:
|
| Click here for a detailed description of this model... |
 "
"