Team:Heidelberg/Notebook/Homology Based/September
From 2010.igem.org
(→01/09/2010) |
(→24/09/2010) |
||
| (8 intermediate revisions not shown) | |||
| Line 121: | Line 121: | ||
==02/09/2010== | ==02/09/2010== | ||
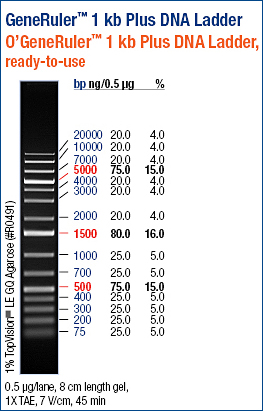
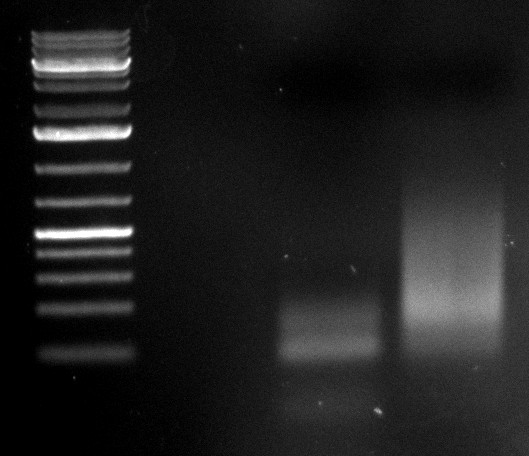
| + | [[Image:Sm133_fam.jpg|200px]] | ||
| + | |||
| + | Ladder description from invitrogen. This ladder was used in all the gels run, in case not labeled, please refer to this image for the information. | ||
*PCR of day before: (1kb Plus) | *PCR of day before: (1kb Plus) | ||
[[Image:20100902_thomas_virshuf_2ndPCR_n.jpg|200px]] | [[Image:20100902_thomas_virshuf_2ndPCR_n.jpg|200px]] | ||
| Line 146: | Line 149: | ||
*nucleotide removal | *nucleotide removal | ||
**c ≈ 37<sup>ng</sup>/<sub>µl</sub> in 32µl each ⇒ enough for 3 PCRs each | **c ≈ 37<sup>ng</sup>/<sub>µl</sub> in 32µl each ⇒ enough for 3 PCRs each | ||
| - | |||
==03/09/2010== | ==03/09/2010== | ||
| Line 387: | Line 389: | ||
==24/09/2010== | ==24/09/2010== | ||
| - | * 5 flasks ( | + | * 5 flasks (125cm2) of HEK cells were transfected with a total of 75 µg of the library DNA and AAV helper plasmid (1:1) for AAV production, using HBSS. However, the transfection was not successful. |
* minipreps of the 50 picked colonies were done, and 10 clones were sent for sequencing. Test digestion with AscI, PacI indicated that the clones picked were positive for the insert (capsid genes). | * minipreps of the 50 picked colonies were done, and 10 clones were sent for sequencing. Test digestion with AscI, PacI indicated that the clones picked were positive for the insert (capsid genes). | ||
| - | * Counting of the colonies on the plates from the previous day showed a library size of | + | * Counting of the colonies on the plates from the previous day showed a library size of 6,4 x 10^7 |
| - | + | ||
| + | * 50 more colonies were picked from one of the plates and used to inoculate mini-prep cultures. | ||
==25/09/2010== | ==25/09/2010== | ||
| Line 408: | Line 410: | ||
* The source of the contamination with AAV5 was identified. The second PCR for the shuffled capsids was repeated. | * The source of the contamination with AAV5 was identified. The second PCR for the shuffled capsids was repeated. | ||
| + | |||
| + | [[Image:2010-09-29 20hr 33min.jpg|thumb|200px|center|2nd PCR for the shuffled cap genes]] | ||
==29/09/2010== | ==29/09/2010== | ||
Latest revision as of 03:16, 28 October 2010

September01/09/2010
500 ng input from purified DNAse digest 10 ul Phusion HF Buffer 1 ul dNTPs 1.5 ul DMSO 0.5 ul Phusion Polymerase ad 50 ul H2O
2 ul Re-Assembly PCR product 1 ul SAFor 1 ul SARev 0.5 ul MgSO4 10 ul 5x Hifi Buffer 33.5 ul H2O 2 ul HiFi Cycles 2-step PCR protocol (Program name: "shuffle phil", left PCR): 95 °C for 5 min 94 °C for 15 sec | 66°C for 30 sec | 40x 68°C for 3 min | 72 °C for 10 min 4 °C hold 02/09/2010Ladder description from invitrogen. This ladder was used in all the gels run, in case not labeled, please refer to this image for the information.
03/09/2010
06/09/2010
07/09/2010
08/09/2010
09/09/2010
10/09/2010
10/09/2010cap Gene Shuffling (starting over again)cap PCRPCR to amplify the cap genes from the stock.
Cycles (Program name: "shuffle 1st"):
Gel Extraction: DNAse Digestcap gene mix (4ug DNA total): B1/B2 (09/15) !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!! AAV m [ng] c [ng/ul] V [ul] 1 350 121.0 2.9 H2O 20.5 DNAse Digest:
Directly after adding of DNAse:
Gel Extraction: Re-Assembly PCR500 ng input from purified DNAse digest 10 ul Phusion HF Buffer 1 ul dNTPs 1.5 ul DMSO 0.5 ul Phusion Polymerase ad 50 ul H2O Amplification PCR2 ul Re-Assembly PCR product 1 ul SAFor 1 ul SARev 0.5 ul MgSO4 10 ul 5x Hifi Buffer 33.5 ul H2O Cycles 2-step PCR protocol (Program name: "shuffle phil", left PCR): 95 °C for 5 min 94 °C for 15 sec | 68°C for 3 min | 40x 72 °C for 10 min 4 °C hold 10/09/2010
ToDo20/09/2010
- 15 µg capsid genes. - 7 µL AscI - 7 µL PacI - 20 µL 10X NEB buffer 4 - 2 µL 100X - Up to 200 µL nuclease-free water
- 10 µg vector - 2 µL AscI - 2 µL PacI - 5 µL 10X BSA - 5 µL 10X buffer 4 - Up to 50 µL nuclease-free water 21/09/2010
- ~3.556 µg insert (Capsid genes) - ~2.667 µg vector - 6 µL T4 DNA ligase - 8 µL T4 DNA ligase buffer - Up to 80 µL nuclease-free water - Incubate at 16 ͦC overnight 22/09/2010
- 600 µL of electrocompetent bacteria were mixed with 62.5 µL DNA from the ligation reaction (about 125 ng per 20 µL bacteria), 20 µL of the mixture were placed in the electoporation cuvette - Electroporation conditions: - Recovery of bacteria was done by adding 1 mL LB media to each 20 µL of bacteria that were electroporated. The bacteria were collected in a 200 mL flask and incubated at 37ͦC with shaking at 225 rpm for 1 hour. The bacteria were then used to inoculate 15 cm petri dishes with ampicillin resistance media. 500 µL were spread on each plate, and 50 plates in total were inoculated. The plates were incubated at 37 ͦC overnight. 23/09/2010
24/09/2010
25/09/2010
27/09/2010
28/09/2010
29/09/2010
30/09/2010
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"