Team:Heidelberg/Notebook/miRNA Kit/October
From 2010.igem.org
(→24/10/2010) |
|||
| (14 intermediate revisions not shown) | |||
| Line 74: | Line 74: | ||
repressor construct: | repressor construct: | ||
* colony 8, 9, 14 and 16 were chosen for miniprep. They consist of the complete TetR construct in pBS 1C3 | * colony 8, 9, 14 and 16 were chosen for miniprep. They consist of the complete TetR construct in pBS 1C3 | ||
| - | * digestion of the minis with EcoRI and PstI for cloning into | + | * digestion of the minis with EcoRI and PstI for cloning into [http://partsregistry.org/Part:pSB1A3 pSB1A3], SAP of [http://partsregistry.org/Part:pSB1A3 pSB1A3], digestion was tested on gel and proven positive (see Gel 101001-1) |
* nucleotide removal, then ligation of TetR8 and TetR14 equimolar with SAPed backbone, TetR9 and TetR16 2:1 with not-SAPed backbone, transformation with TOP10 cells (10µl on 50µl ''E. coli'') | * nucleotide removal, then ligation of TetR8 and TetR14 equimolar with SAPed backbone, TetR9 and TetR16 2:1 with not-SAPed backbone, transformation with TOP10 cells (10µl on 50µl ''E. coli'') | ||
<br /> | <br /> | ||
| Line 94: | Line 94: | ||
repressor construct:<br/> | repressor construct:<br/> | ||
| - | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of | + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of previous day transformations |
** analysis, see gel 101002-1 | ** analysis, see gel 101002-1 | ||
* twice test digestion of promising mini-preps, following [https://2010.igem.org/3A_Assembly standard protocol recommendations] <br/> | * twice test digestion of promising mini-preps, following [https://2010.igem.org/3A_Assembly standard protocol recommendations] <br/> | ||
| Line 122: | Line 122: | ||
===04/10/2010=== | ===04/10/2010=== | ||
repressor construct:<br/> | repressor construct:<br/> | ||
| - | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of | + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of 6 colonies for each plate of previous day transformations |
** gel 101004-1 reveals almost positive samples | ** gel 101004-1 reveals almost positive samples | ||
* submission of each a promising mini-prep for sequencing of binding site and correct insert | * submission of each a promising mini-prep for sequencing of binding site and correct insert | ||
| Line 128: | Line 128: | ||
===05/10/2010=== | ===05/10/2010=== | ||
repressor construct:<br/> | repressor construct:<br/> | ||
| - | * <i>sequencing results</i> from | + | * <i>sequencing results</i> from previous day: |
** once or twice the binding site in each of the T1 constructs, respectively | ** once or twice the binding site in each of the T1 constructs, respectively | ||
** once the binding site for the T2 construct | ** once the binding site for the T2 construct | ||
*** re-trial of ligation for two binding sites in T2 | *** re-trial of ligation for two binding sites in T2 | ||
| - | * planned transfection into HeLa and HUH cells (non-liver and liver to prove ON-targeting) in ratio 6:1 = repressor:operator(tuning) as recommended in the [http:// | + | * planned transfection into HeLa and HUH cells (non-liver and liver to prove ON-targeting) in ratio 6:1 = repressor:operator(tuning) as recommended in the [http://www.roche-applied-science.com/pack-insert/1815091a.pdf manual] |
new approach: | new approach: | ||
| Line 141: | Line 141: | ||
===06/10/2010=== | ===06/10/2010=== | ||
| - | * waiting for colonies from | + | * waiting for colonies from previous night, which did not grow after all |
===07/10/2010=== | ===07/10/2010=== | ||
* repetition of shhAAT cloning | * repetition of shhAAT cloning | ||
* touch-down PCR setup: 8.5 µl ddH<sub>2</sub>O, 0.5 µl template DNA (pcDNA5, 50 ng/µl), 0.5 µl each primer (i.e. [[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D7 & D16]], 10 µl Phusion PCR MasterMix (2x) | * touch-down PCR setup: 8.5 µl ddH<sub>2</sub>O, 0.5 µl template DNA (pcDNA5, 50 ng/µl), 0.5 µl each primer (i.e. [[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D7 & D16]], 10 µl Phusion PCR MasterMix (2x) | ||
| - | * touch-down PCR protocol: [ | + | * touch-down PCR protocol: [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Plasmid-PCR like this] but with increase of annealing temperature from 68°C to 61°C (i. e. 0.2°C per cycle) |
* go on like [[Igem2010/Main/synthetic miR Kit/October#05/10/2010 | 05/10/2010]] but growth on freshly prepared plates | * go on like [[Igem2010/Main/synthetic miR Kit/October#05/10/2010 | 05/10/2010]] but growth on freshly prepared plates | ||
| Line 153: | Line 153: | ||
===09/10/2010=== | ===09/10/2010=== | ||
| - | * sequencing | + | * sequencing the day before was fine but construct contains TetO<sub>2</sub>, thus: maybe Q2 and Q3 were confused |
** that would explain why measurements with TetR constructs are not working properly even though sequences were fine | ** that would explain why measurements with TetR constructs are not working properly even though sequences were fine | ||
* cloning of binding sites for shhAAT (perfect binding site generated via annealing, and randomized ones PCR amplified) | * cloning of binding sites for shhAAT (perfect binding site generated via annealing, and randomized ones PCR amplified) | ||
** touch-down PCR setup: 17 µl ddH<sub>2</sub>O, 4 µl each oligo (i.e. respective forward primer + second strand oligo[[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D121]]), 25 µl Phusion PCR MasterMix (2x) | ** touch-down PCR setup: 17 µl ddH<sub>2</sub>O, 4 µl each oligo (i.e. respective forward primer + second strand oligo[[Igem2010/Main/Primer_database#Standart_Kit_Cloning_Primers | D121]]), 25 µl Phusion PCR MasterMix (2x) | ||
| - | *** touch-down PCR protocol: [ | + | *** touch-down PCR protocol: [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#shRNA-PCR_protocol like this] but with increase of annealing temperature from 70°C to 62°C (i. e. 0.5°C per cycle, 16x) and four additional cycles with 62°C |
** annealing of perfect binding against shhAAT sites with an oligo-based strategy following the protocol from the [[Igem2010/Main/synthetic miR Kit/October#03/10/2010 | 3<sup>rd</sup> October]] | ** annealing of perfect binding against shhAAT sites with an oligo-based strategy following the protocol from the [[Igem2010/Main/synthetic miR Kit/October#03/10/2010 | 3<sup>rd</sup> October]] | ||
** digestion of PCR amplified binding sites with AgeI and XhoI, digestion of Q2 backbone containing the shhAAT with XhoI and XmaI, afterwards heat inactivation, nucleotide removal and ligation following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ** digestion of PCR amplified binding sites with AgeI and XhoI, digestion of Q2 backbone containing the shhAAT with XhoI and XmaI, afterwards heat inactivation, nucleotide removal and ligation following [https://2010.igem.org/3A_Assembly standard protocol recommendations] | ||
| Line 164: | Line 164: | ||
===10/10/2010=== | ===10/10/2010=== | ||
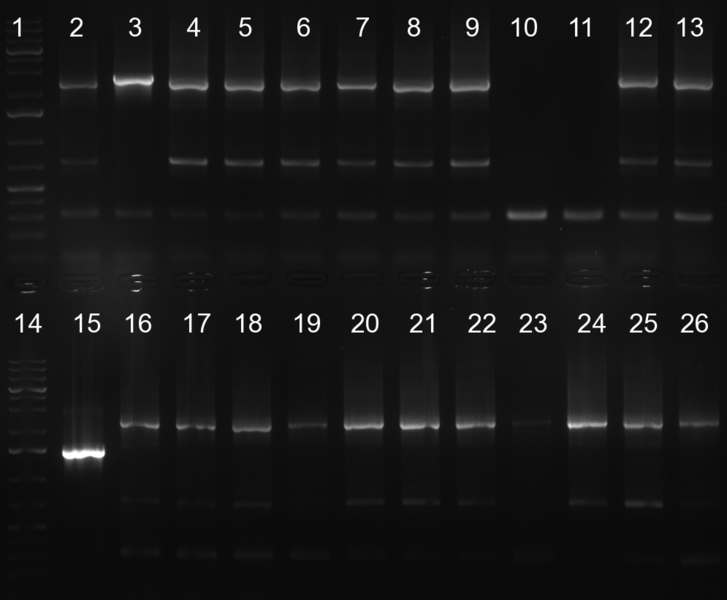
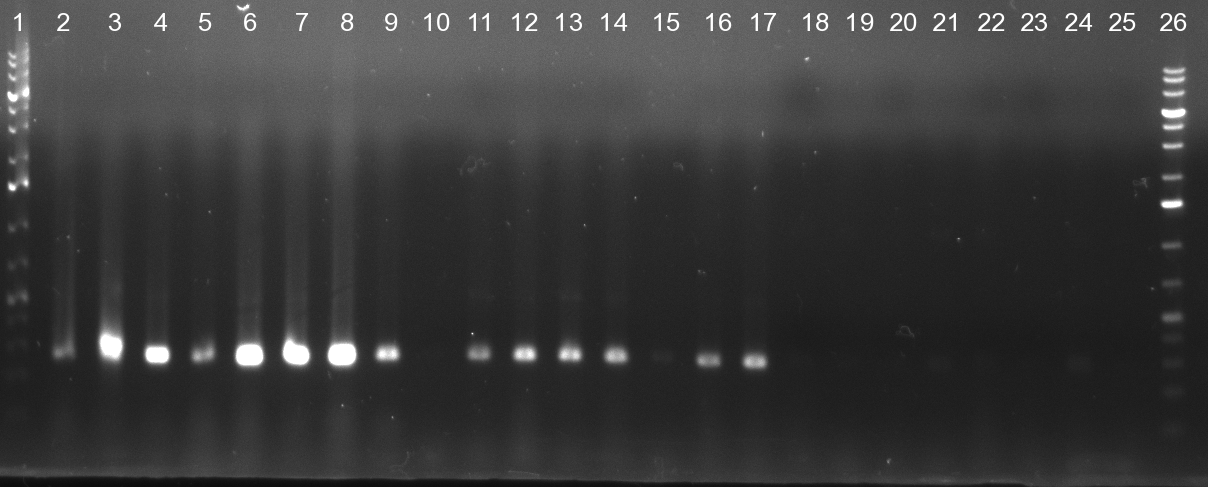
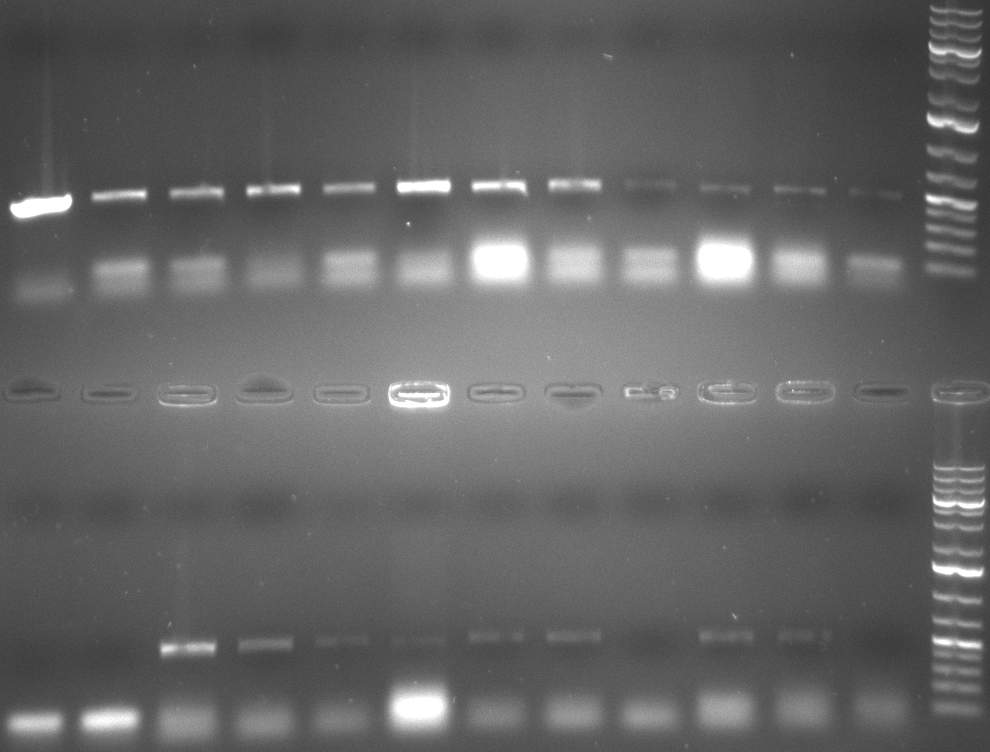
[[Image:20101010 colonypcr shhAAT p 12 22 bs LOLA.png|thumb|350px|right|Gel 101010-1. Colony PCR for construct containing shhAAT and binding sites. Expected is an amplified fragment a bit smaller than 300 bp which can be seen on lane 3, 4, 6, 7, 8 (perfect binding sites), 12, 13, 14, 16 and 17 (randomized binding sites). Lane 1 and Lane 26: 1kb Plus Ladder (Invitrogen)]] | [[Image:20101010 colonypcr shhAAT p 12 22 bs LOLA.png|thumb|350px|right|Gel 101010-1. Colony PCR for construct containing shhAAT and binding sites. Expected is an amplified fragment a bit smaller than 300 bp which can be seen on lane 3, 4, 6, 7, 8 (perfect binding sites), 12, 13, 14, 16 and 17 (randomized binding sites). Lane 1 and Lane 26: 1kb Plus Ladder (Invitrogen)]] | ||
| - | * colony pcr reveals some positive clones (see gel 101010-1) from | + | * colony pcr reveals some positive clones (see gel 101010-1) from previous day |
** inoculation of 5 ml LB Amp cultures for mini-prep over night | ** inoculation of 5 ml LB Amp cultures for mini-prep over night | ||
| Line 182: | Line 182: | ||
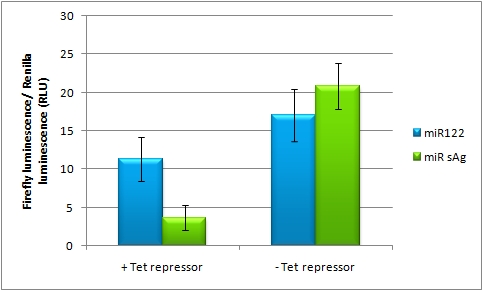
In presence of Tet repressor and miR122 that is not targeting it, less of firefly luciferase is expressed, but when miR sAg is expressed is recovered to 60% of original activity. | In presence of Tet repressor and miR122 that is not targeting it, less of firefly luciferase is expressed, but when miR sAg is expressed is recovered to 60% of original activity. | ||
| + | <br /><br /> | ||
| + | <br /><br /> | ||
| + | <br /><br /> | ||
| + | <br /><br /> | ||
| + | <br /><br /> | ||
===11/10/2010=== | ===11/10/2010=== | ||
| - | * mini-prep of ten cultures from | + | * mini-prep of ten cultures from previous day and direct transfection to test the system |
** amount: each 100 µl (c=2.5 ng/µl) | ** amount: each 100 µl (c=2.5 ng/µl) | ||
<br/> | <br/> | ||
| Line 195: | Line 200: | ||
*** 25 µl Phusion PCR MasterMix (2x) | *** 25 µl Phusion PCR MasterMix (2x) | ||
** touch-down PCR protocol: | ** touch-down PCR protocol: | ||
| - | *** [ | + | *** [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#shRNA-PCR_protocol like this] but with increase of annealing temperature from 70°C to 62°C (i. e. 0.5°C per cycle, 16x) |
*** and four additional cycles with 62°C, elongation time only 10 seconds | *** and four additional cycles with 62°C, elongation time only 10 seconds | ||
* digestion of generated binding sites and the pSMB_miMeasure backbone with EcoRI and PstI, then nucleotide removal | * digestion of generated binding sites and the pSMB_miMeasure backbone with EcoRI and PstI, then nucleotide removal | ||
| Line 207: | Line 212: | ||
*** elongation time: 90<nowiki>''</nowiki> | *** elongation time: 90<nowiki>''</nowiki> | ||
** <u>no positive</u> results, maybe due to insufficient screening primers (one for BBb standard, the other one for CMV resulting in an 1300 bp fragment) | ** <u>no positive</u> results, maybe due to insufficient screening primers (one for BBb standard, the other one for CMV resulting in an 1300 bp fragment) | ||
| - | *** as a consequence: repetition of entire cloning with [http://www.fermentas.com/templates/files/tiny_mce/coa_pdf/coa_ef0511.pdf SAP] treated backbone | + | *** as a consequence: repetition of entire cloning with [http://www.fermentas.com/templates/files/tiny_mce/coa_pdf/coa_ef0511.pdf SAP] treated backbone following day |
===13/10/2010=== | ===13/10/2010=== | ||
measurement construct for single binding sites: <br/> | measurement construct for single binding sites: <br/> | ||
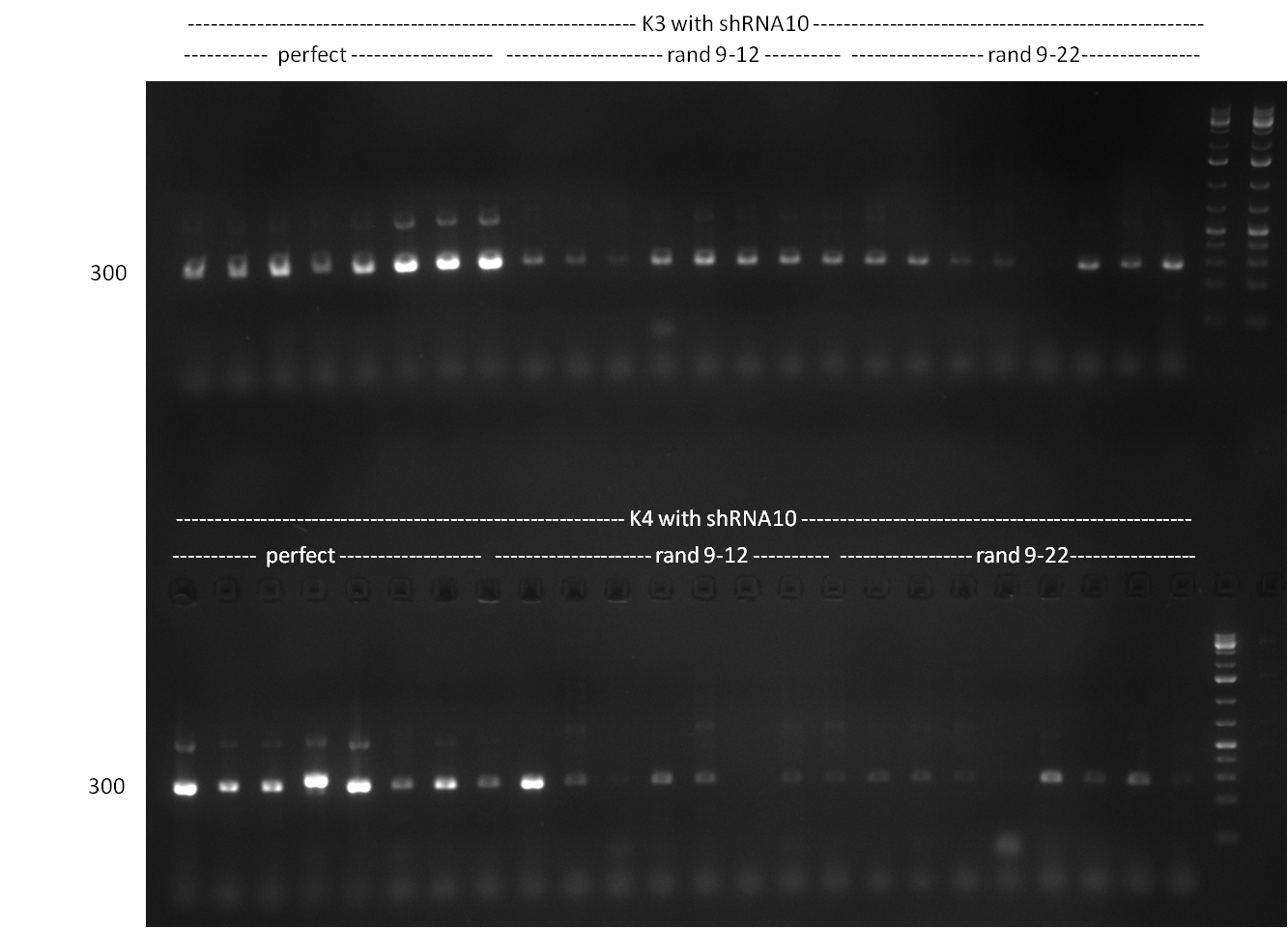
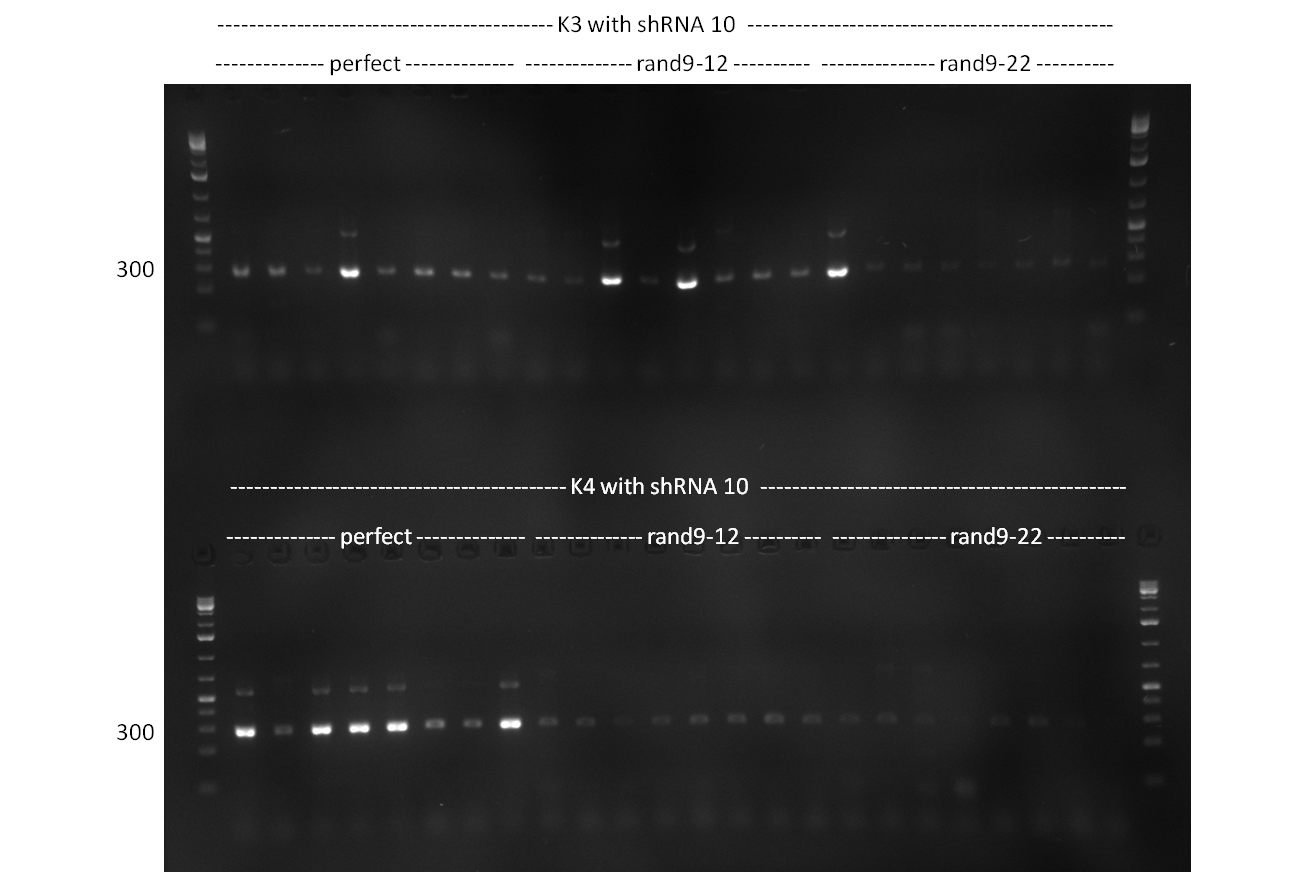
| - | * repetition of cloning from | + | * repetition of cloning from 11/10/2010 |
* <u>note:</u> vector concentration of pSMB_miMeasure is okay but after digestion and nucleotide removal it is always pretty low | * <u>note:</u> vector concentration of pSMB_miMeasure is okay but after digestion and nucleotide removal it is always pretty low | ||
** (loss of 75% of DNA on average whereas other DNA yields to 90% of purified DNA as compared to digested amount) | ** (loss of 75% of DNA on average whereas other DNA yields to 90% of purified DNA as compared to digested amount) | ||
| Line 259: | Line 264: | ||
===18/10/2010=== | ===18/10/2010=== | ||
| - | * [ | + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Colony_PCR colony-PCR] of 4 colonies for each plate of [[Igem2010/Main/synthetic miR Kit/October#17/10/2010 | previous day transformations]] using reverse binding site oligo and forward hAAT primer expecting a fragment round about 1300 bp |
** gel 101018-1 reveals almost positive samples except two (data not yet shown) | ** gel 101018-1 reveals almost positive samples except two (data not yet shown) | ||
| Line 265: | Line 270: | ||
* mini-preps of cloned shAAT constructs | * mini-preps of cloned shAAT constructs | ||
* dilution and co-transfection with miRsAg expressing construct into HeLa cells for ELISA measurements | * dilution and co-transfection with miRsAg expressing construct into HeLa cells for ELISA measurements | ||
| + | |||
| + | ===23/10/2010=== | ||
| + | seeding HeLa cells | ||
| + | |||
| + | ===24/10/2010=== | ||
| + | [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Transfection Transfection] | ||
| + | |||
| + | 1) HeLa luc2-miR122BS and miR122 | ||
| + | 2) for ELISA - pBS_U6 containing hAAT with miRsAg imperfect binding sites, and miRsAg | ||
| + | ratios 1:1 ans 4:1 | ||
| + | |||
| + | ===26/10/2010=== | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay] | ||
| + | |||
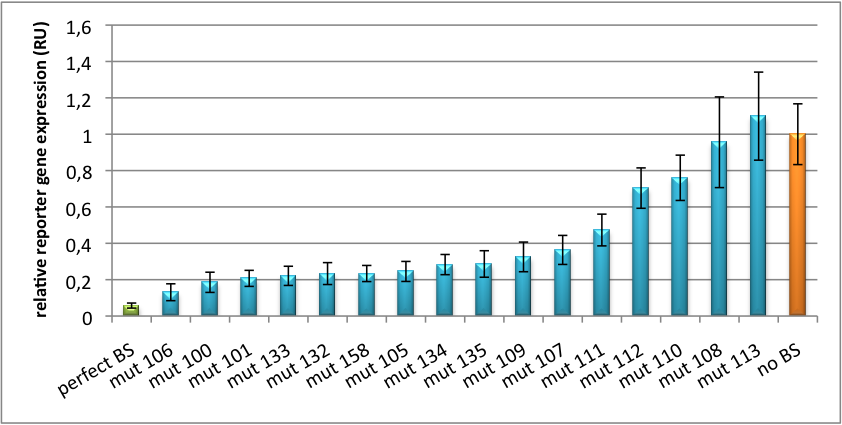
| + | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
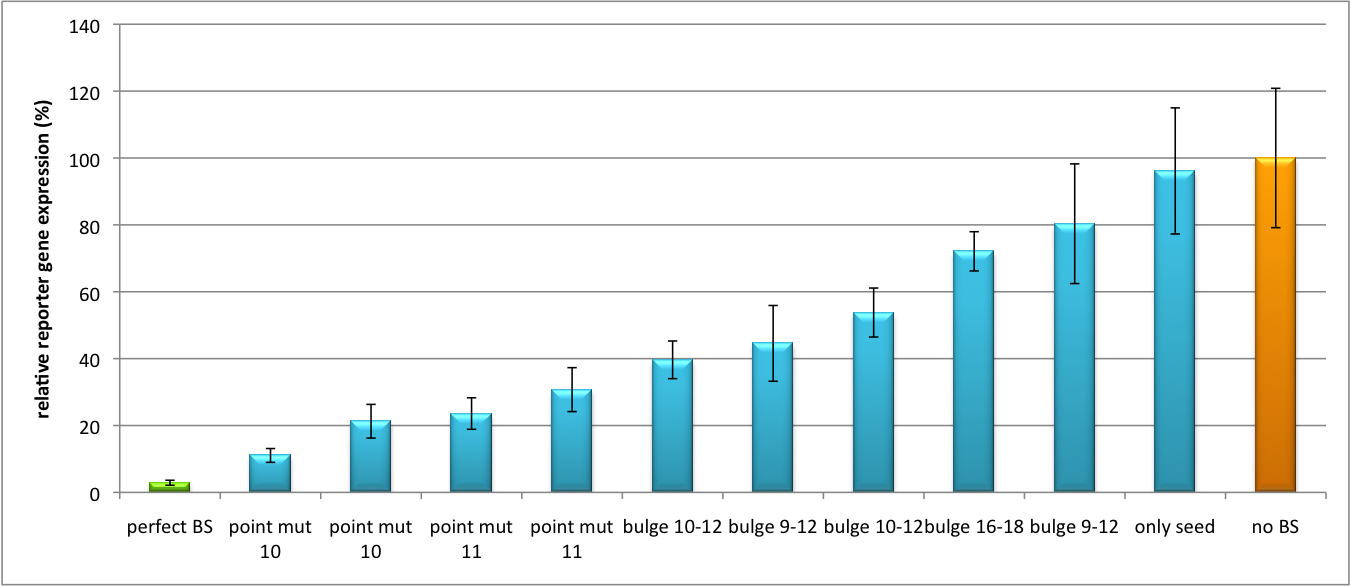
| + | [[Image:Haat U6HD2010.jpg|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
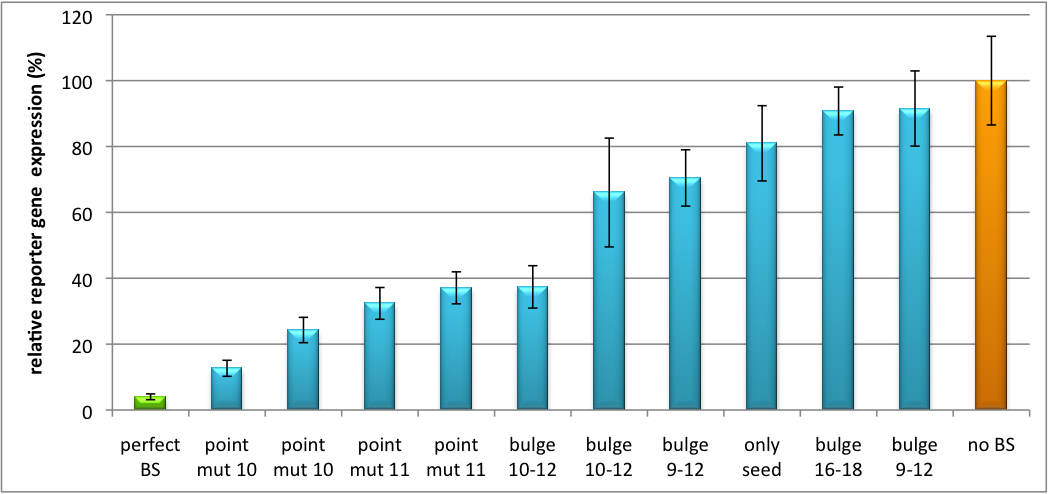
| + | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
| + | <br><br><br> | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] was performed on media in which HeLa cells we grown, as hAAT is secreted by cells. | ||
{{:Team:Heidelberg/Single_Bottom}} | {{:Team:Heidelberg/Single_Bottom}} | ||
Latest revision as of 09:58, 27 October 2010

01/10/2010repressor construct:
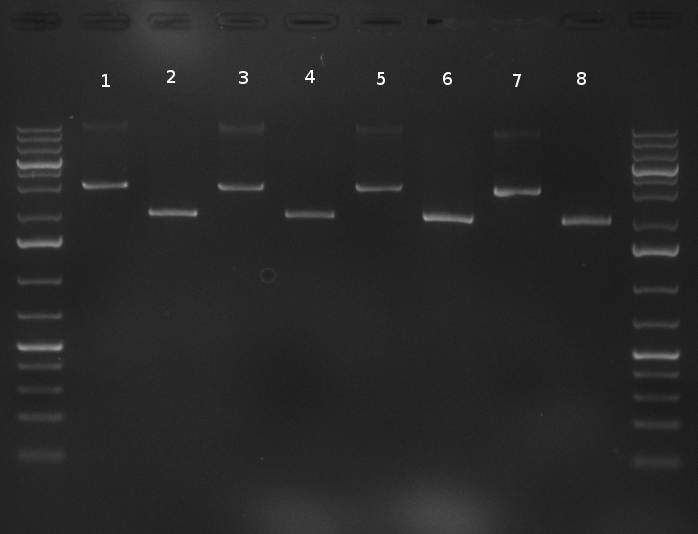
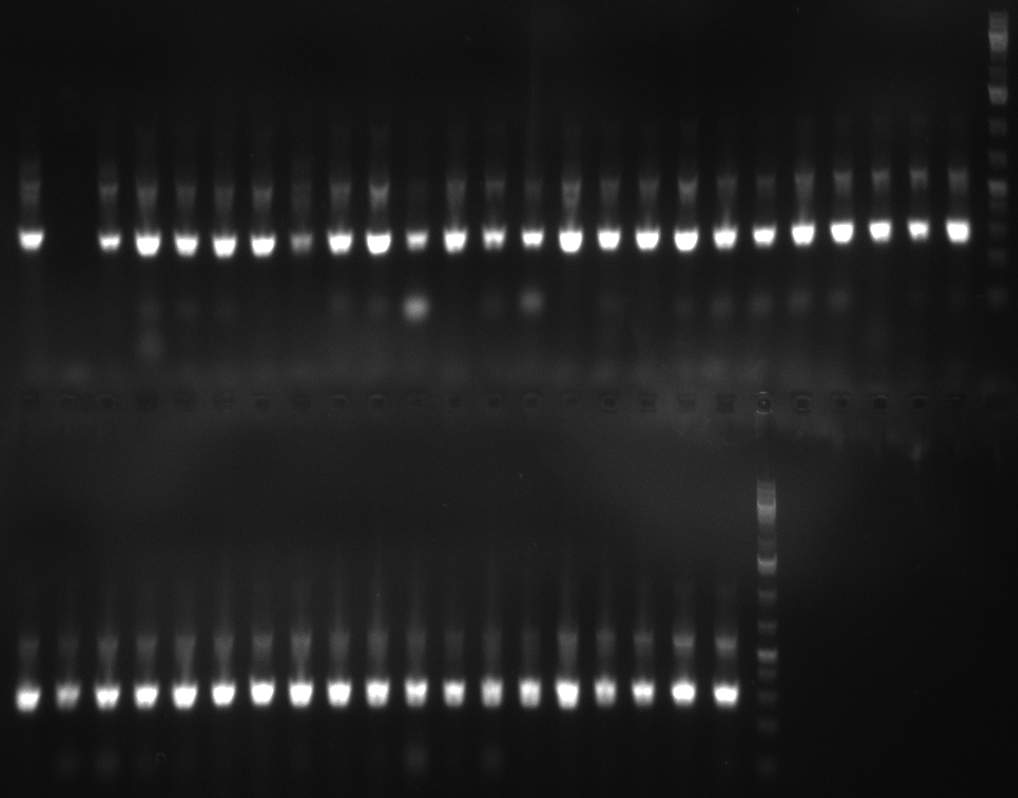
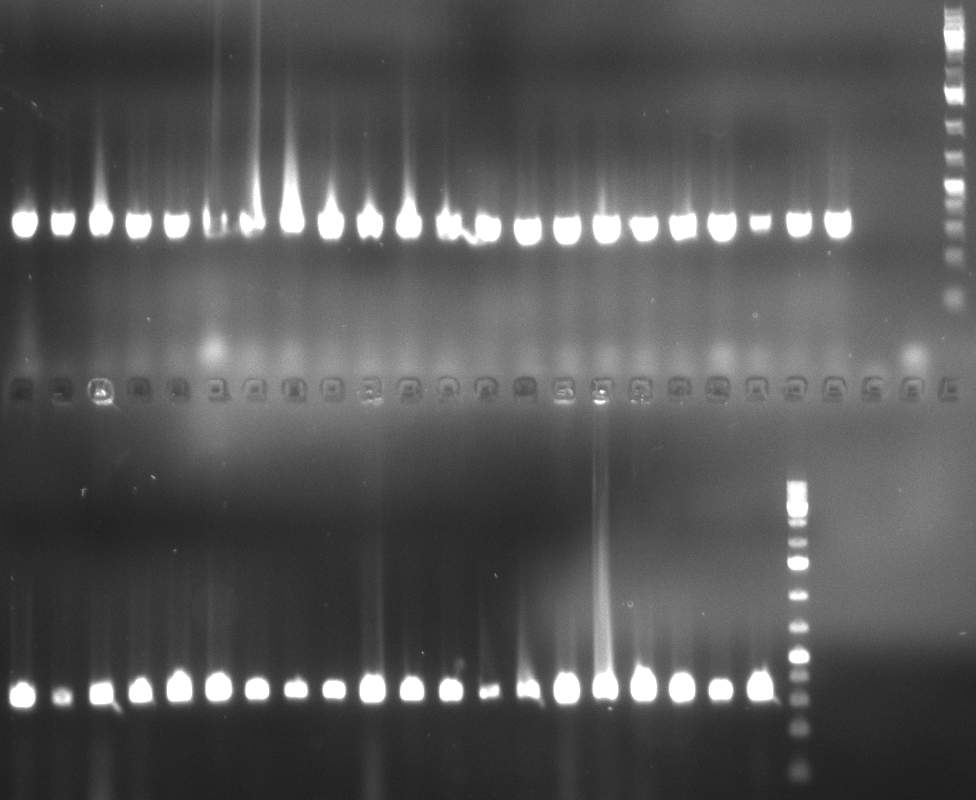
02/10/2010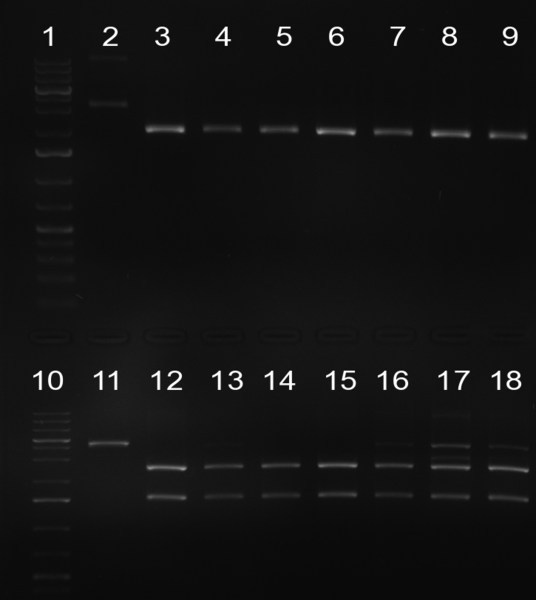 Gel 101002-2. Test digestions of repressor construct. Upper pattern: EcoRI & PstI. Lower pattern: BamHI. All bands revealing positive results except in the first column (negative control from lane 3 of gel 101002-1). Lane 1 and Lane 10: 1kb Plus Ladder (Invitrogen). Lane 16, 17, 18: incomplete digestion
03/10/2010repressor construct:
04/10/2010repressor construct:
05/10/2010repressor construct:
new approach:
06/10/2010
07/10/2010
08/10/2010
09/10/2010
10/10/2010
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 Tuning construct in which luc2 expression is driven by CMV promoter with TetO2] also expressing either miR122 or miR sAg was cotransfected with Tet repressor under regulation of perfect miR sAg binding site. Amounts transfected were: 2.5ng of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 tuning construct], 20ng of miRNA expressing plamid (either miR122 or miR sAg) and 5ng of repressor construct. To keep transfection efficiency the same we used sheared salmon sperm DNA as a stuffer.
11/10/2010
measurement construct for single binding sites:
12/10/2010measurement construct for single binding sites:
13/10/2010measurement construct for single binding sites:
14/10/2010
15/10/2010
16/10/2010
17/10/2010
18/10/2010
19/10/2010
23/10/2010seeding HeLa cells 24/10/20101) HeLa luc2-miR122BS and miR122 2) for ELISA - pBS_U6 containing hAAT with miRsAg imperfect binding sites, and miRsAg ratios 1:1 ans 4:1 26/10/2010
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"