Team:Edinburgh/Notebook/Red light producer
From 2010.igem.org
Macbereska (Talk | contribs) (→Red Light Producer) |
|||
| (6 intermediate revisions not shown) | |||
| Line 43: | Line 43: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 50: | Line 50: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the project</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | ||
| Line 59: | Line 59: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 72: | Line 72: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 79: | Line 79: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Branding">iGEM survey</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/FutureApps">future applications</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Human">further thoughts</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 125: | Line 117: | ||
<div id="windowbox" style="border: .2em solid #660000; padding: 5px; position:fixed; top:50%; right:30px; width:8%;"> | <div id="windowbox" style="border: .2em solid #660000; padding: 5px; position:fixed; top:50%; right:30px; width:8%;"> | ||
| - | <span style="color:ivory;"> | + | <span style="color:ivory;">These <b>notes</b> correspond to the part of the <b>project</b> detailed <a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">here</a>.</span> |
</div> | </div> | ||
| Line 301: | Line 293: | ||
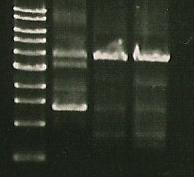
*Gel of the ligations: marker (lane 1), 356K (lane 2,3), S284T (lane 3,4), 358Rs (lane 5,6). | *Gel of the ligations: marker (lane 1), 356K (lane 2,3), S284T (lane 3,4), 358Rs (lane 5,6). | ||
| - | [Image:163.jpg]] | + | [[Image:163.jpg]] |
'''25/8/2010''' | '''25/8/2010''' | ||
| Line 308: | Line 300: | ||
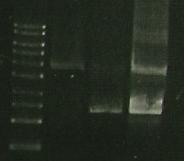
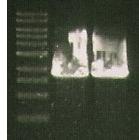
*Did MABEL PCR PPyInc + S284T primers | *Did MABEL PCR PPyInc + S284T primers | ||
*used 0.5 ul template DNA | *used 0.5 ul template DNA | ||
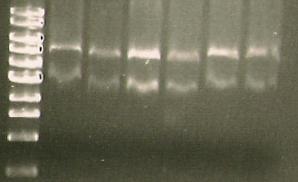
| + | *Gel: marker (Lane1), ligation of PCR product for 356R (Lane2), Ligation of 07 PCR for S284T (lane3), Pure PCR from S284T. | ||
| + | |||
| + | [[Image:164.jpg]] | ||
'''26/8/2010''' | '''26/8/2010''' | ||
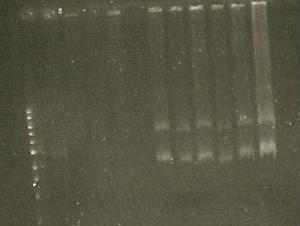
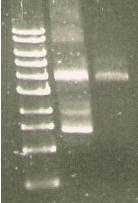
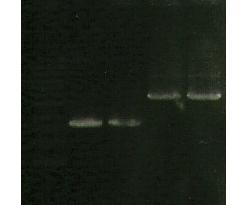
| Line 317: | Line 312: | ||
** N 99 G | ** N 99 G | ||
** SKL- IAV | ** SKL- IAV | ||
| + | |||
| + | [[Image:165.jpg]] | ||
'''27/8/2010''' | '''27/8/2010''' | ||
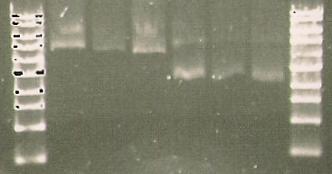
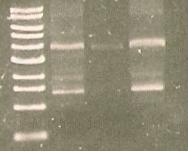
| Line 368: | Line 365: | ||
* mutation of S284T (right) | * mutation of S284T (right) | ||
| + | [[Image:168.jpg]] | ||
'''9/9/2010''' | '''9/9/2010''' | ||
| + | |||
| + | [[Image:170.jpg]] | ||
| + | |||
{| border="1" | {| border="1" | ||
| Lane 1 | | Lane 1 | ||
Latest revision as of 02:11, 28 October 2010
Red Light Producer
29/6/10
Now preparing primers to make a set of different spectra bioluminescent proteins. Need to order primers for Site directed mutagenesis of the Firely luciferase to make it red (MABEL protocol), and to make click beetle luciferase Rluc8 (which emits light at about 420nm in presence of bisdeoxycoelenterazine - about the same price a coelenterazine though)
8/7/2010
- Red luciferase mutagenesis primers arrived- 3 sets:
- S248T
- 356 K
- 356 R
- Followed the protocol with 34ul water + 0.5 template DNA
- Put the primers and template in green box (???)
- Ran Gel: 5 µL PCR product, 5 µL water, 2.5 µL buffer
| Marker | S284T | 356K | 356R |
- 356K and 356R look fine
- going to run S284T at a higher ** temp tomorrow to see if it works better
- all PCR tubes stored in freezer
09/7/10
Redo PCR of Red Luciferase
- Everything except template DNA + Kod polymerase kept on ice
- Ran PCR
- Template DNA used PyeaR and P. pyralis luciferase BBa_K216015
- Purification done for 356K and 356R; stored in iGEM box (-20C).
9/7/2010
- Ligations set up overnight- might have put too much ligase in 356K... Incubated at 15C.
- Oure 356R and 356K are in the freezer- row 10 (G &H)?
13/07/10 Goal of the day:
Transformations:
- 356R (2): one from 16 °C and one from room temp(RT)
- 356K (2): one from 16 °C and one from room temp(RT)
- S284T (2): one with 25/5 ligase, one with 1/7 ligase
- I had a burger for the lunch. It was very nice.
- Now we are transforming cells:
** 356R (2)& 356K (2): one form 16C, one from RT ** S248T (2): 1 with 25/5 ligase, other with 1.7.10 ligase/
Empty edinbrick vector for control.
- LuxAB + edinbrick ligation.
- Using 100 ul transformed cells + 800 ul LB for all. Incubation started half 5.
22/07/10
Liquid Cultures (amp 80):
- 356R (RT) – 1 with NaNO3 and 1 without
- (16°C) – 2 with NaNO3 and 2 without
- 356K (RT) – 1 with NaNO3 and 1 without
- (16°C) – 2 with NaNO3 and 2 without
- S284T (100 µL and 25/5 ligase) – 2 with NaNO3 and 2 without
- (900 µL and 25/5 ligase) – 3 with NaNO3 and 3 without
- (900 µL and 1/7 ligase) – 5 with NaNO3 and 5 without
NaNO3 = 30mM Sodium Nitrate
23/07/10
Double digest of K098010
28/07/10
Primer design to produce a stable red light luciferase. The mutations for increasing bioluminescence come from [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W9V-4NHM57C-1&_user=809099&_coverDate=07%2F15%2F2007&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1414502247&_rerunOrigin=google&_acct=C000043939&_version=1&_urlVersion=0&_userid=809099&md5=00d9c92f6452bd276c6ea606ed7d3bbf Fujii et al, 2007] and the paper from [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W9V-4X6MSYB-7&_user=809099&_coverDate=01%2F15%2F2010&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1414503636&_rerunOrigin=google&_acct=C000043939&_version=1&_urlVersion=0&_userid=809099&md5=297be52832193b4d026953632a1edf3c Branchini et al, 2010]
29/7/2010
- Double digest with EcoRI HF&PstI in buffer 2 (neb).
- 2 bands: with and without vector?
- did not leave the digest for too long...
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 |
| Ladder | S284T (4) | S284T (6) | 356R (6, 900) | 356R (6) | 356K (4) |
2/8/2010
- Red luciferases EcoRI HF&PstI double digests- lanes 2-7 in the second miniprep set.
- Ladder didn't work too well ,though...
- Transform last years luciferase
- Look at papers for expression in E. coli
- Replace B0034 by stronger RBS?
- Confirm that we order bright luc.
- Single digest of constructs
- Test glowing with positive control
9/8/2010
Digest:
- WT1
- 356 R: (2)
- 356 K: (4)
- S284T: (6)
- 3 eith EcoRI; 3 with EcoRi + PstI
10/8/2010
- Sequencing of red mutant luciferases.
- Forward primers for S284T and reverse for 356R and K should do.
- Concentration of primers for sequencing: 5uM.
- sequencing tubE: 6 ul liquid: 3ul DNA + 2ul water, 1 ul primer mix.
- when using miniprep DNA- remember to spin down the dirt- 1min, max speed, then take the DNA from the top to avoid disturbing any dirt that is settled.
12/8/2010
- Got the sequences for red luciferases
- see: if we can detect Red Light with the luminometer?
- Red Luciferases 356R and S284T were nNOT MUTANTS- just WT
- set up ;iquid cultures with 30 ul nitrate for:
- 356R - no 3,1 from RT plate, no 5 from 16C plate; no 1 from Master plate
- S284T - 5 from masterplate; 2, 4, 6 from masterplate
- 356K: 4 from masterplate
13/8/2010 Big day today... TESTING YESTERDAYS PLAN- LUMINESCENCE
16/8/2010
- Sequecning of:
- WR1- 356R -use S284T forward primer
- WR2 -S284T -use 356 R reverse primer
- WR3- LovTap 03
- WR4 -LovTap 09
- RESULTS: ONLY 356K WAS A REAL MUTANT; S284T AND 356R TURNED OUT TO BE STILL A WT
19/8/2010
- star6: 56,869
- star4: 13,471
- 356K: 263,778
- star 2: 3,717
- WT: 1,617832
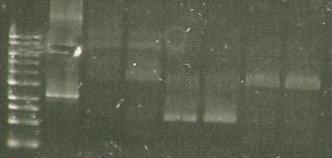
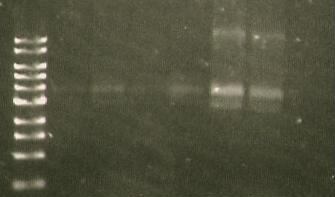
- Gel of the ligations: marker (lane 1), 356K (lane 2,3), S284T (lane 3,4), 358Rs (lane 5,6).
25/8/2010
- Next step:- redo ligations for 356R; redo PCR for S284T
- Ligations
- Did MABEL PCR PPyInc + S284T primers
- used 0.5 ul template DNA
- Gel: marker (Lane1), ligation of PCR product for 356R (Lane2), Ligation of 07 PCR for S284T (lane3), Pure PCR from S284T.
26/8/2010
- check the Fuji et. al paper, see what sequence they used to start off with.
- check IAV in other luciferase
- Fix scar before RBS and in teh second stop codon.
- compared to WT, luciferase sequence has:
- N 50 P
- N 99 G
- SKL- IAV
27/8/2010
- Ligations from yesterday out of freezer.
- Running on gel
- Column 3 9with 356R) has less DNA in it- not enough from ligation and messed up loading of gel).
| Lane 1 | Lane 2 | Lane 3 | Lane 4 |
| Ladder | New S284T PCR - new ligase and the buffer | 356 R ligation | S284T PCR + old ligase |
30/8/2010
- Primers have arrived for fixing the luciferase- removing 6 bases
- Primers have arrived for up and downstream sequences of TnaA region.
- Send Chairman sequence for T7 promoter
- transformed 356R and S284T ligations from Friday
- Used PCR product as a control
- Plated out the transformed cells. Used 900 rather than 100 if possible
31/8/2010
- We have S284T transformants but
- 14 colonies on ligation plate
- 4 colonies on control where we used PCR product instead of ligation
- For sequencing-
- if sequencing a S284T mutant, use 356R/K reverse primer
- if sequencing 356R mutant, use S284T forward primer.
2/9/2010
- minipreped 14 colonies (1-14)
- run the gel with samples digested with EcoRI
- here: samples 1-7
3/9/2010
- S284T 8-14 (from yesterday)
8/9/2010
- wWT of luciferase (left)
- mutation of S284T (right)
9/9/2010
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 |
| Ladder | WT | S284T 1 | luxCDE 9:4 | LuxCDE 9:1 |
20/9/2010
- send for sequencing: 356K, S284T, WT (samples from 16/9: WT- 900, 2; also 16/9 S284T- 900, 4.)
- all in psB1C3 vector.
- Transform 356K and S284T with RBS and promoter (lacZ, LacI, RBS).
- Run gel of 365K in psb1C3 of the digest. --> control: RFP + psb1C3
- then add promoter
23/9/2010
- Added in incubator room liquid culture ?) LacP + RBS + Luciferase for S284T, 356K and WT.
- miniprep- 6 bottles
- GLOWING
- S248T-
- 9.2; 5
- 9.4; 1
- 5; 4
- 356
- 3.3
- WT
- 3.2; 4
- 1.4, 5
 "
"