Team:Uppsala-SwedenWeek13
From 2010.igem.org
(→Week-13) |
(→Construction Of J1,J2,J3) |
||
| (5 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
| - | == | + | == Construction Of J1,J2,J3 == |
| - | |||
| - | + | Plasmid construction and verification of lengths for the last two levels, use of specific restriction sites to re-verify the constructs. Colonies obtained in the cells from the previous step were picked from the plate. Each colony was placed in 10ul of LB | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | 1ul of this LB was used to perform colony PCR. | |
| - | + | The product that obtained after running colony PCR were run on gel and plasmids lengths were verified. Colonies with correct plasmid lengths were selected and inoculated using the remaining 9ul of LB. Plasmid of the samples which showed right size band on the gel were sent to a plasmid concentration test. We decided only J1, J2, J3 were in good concentration to proceed. | |
| - | J1, J2, J3 | + | |
| - | + | The inoclum obtained from the overnight culture was used for plasmid extraction. The extracted plasmids were cut at specific restriction sites present in the biobrick sequence and run on the gel. | |
| + | |||
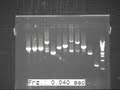
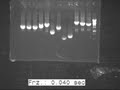
| + | However, we got the correct PCR lengths for Bio-bricks and results are shown in image files below. | ||
| + | The result was showed in Fig.1 and Fig2 | ||
| + | |||
| + | |||
| + | [[Image:20100827 J1-J3 c-PCR 20100826 2 thumb.jpg|500px|center|border|thumb|J1-J3 Figure 1]] | ||
| + | |||
| + | |||
| + | [[Image:20100827 J2-J3 c-PCR 20100826 2 thumb.jpg|500px|center|border|thumb|J2-J3 Figure 2]] | ||
| - | |||
| - | |||
| - | The | + | We got the correct band lengths for all J1, J2, J3. And the one that are with correct band lengths were |
| + | selected. The correct length samples of J1, J2, J3 were again inoculated again to get the enough plasmid for further progress. | ||
Latest revision as of 20:34, 26 October 2010



Week-13
Construction Of J1,J2,J3
Plasmid construction and verification of lengths for the last two levels, use of specific restriction sites to re-verify the constructs. Colonies obtained in the cells from the previous step were picked from the plate. Each colony was placed in 10ul of LB
1ul of this LB was used to perform colony PCR.
The product that obtained after running colony PCR were run on gel and plasmids lengths were verified. Colonies with correct plasmid lengths were selected and inoculated using the remaining 9ul of LB. Plasmid of the samples which showed right size band on the gel were sent to a plasmid concentration test. We decided only J1, J2, J3 were in good concentration to proceed.
The inoclum obtained from the overnight culture was used for plasmid extraction. The extracted plasmids were cut at specific restriction sites present in the biobrick sequence and run on the gel.
However, we got the correct PCR lengths for Bio-bricks and results are shown in image files below. The result was showed in Fig.1 and Fig2
We got the correct band lengths for all J1, J2, J3. And the one that are with correct band lengths were
selected. The correct length samples of J1, J2, J3 were again inoculated again to get the enough plasmid for further progress.
 "
"