Team:Heidelberg/Notebook/Methods
From 2010.igem.org
(→HBSS) |
(→Microscopy) |
||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Pagetop|note_methods}} |
| + | {{:Team:Heidelberg/Side_Top}} | ||
| + | |||
| + | __TOC__ | ||
| + | |||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
=Methods= | =Methods= | ||
==Cloning== | ==Cloning== | ||
| - | === | + | ===Agarose Gel Electrophoresis=== |
| - | + | Agarose flat-bed gels in various concentrations (0.6–2% agarose in 0.5 x TAE buffer) and sizes were run to separate DNA fragments in an electrical field (10–20 V/cm) for analytical or preparative use. The desired amount of agarose was boiled in 1 x TAE buffer until it was completely dissolved. After it cooled down to approximately 60°C, ethidium bromide (EtBr) solution (0.5 µg/ml final concentration) was added to the liquid agar, which was then poured in a flat-bed tray with combs. As soon as the agarose solidified, the Running buffer (0.5 x TAE buffer) was added before the DNA in the loading buffer was loaded into the wells and separated electrophoretically. Ethidium bromide intercalates with the DNA’s GC ntss resulting in DNA-EtBr-complex that emits visible light. Therefore, the DNA fragments could be detected on a UV-light tray at 265 nm. | |
<br><br> | <br><br> | ||
| - | === | + | ===Colony PCR=== |
| - | performed by | + | |
| + | Colony PCRs were performed using Fermentas PCR Master Mix (2x), containing ''Taq'' DNA Polymerase.<br/> | ||
| + | Bacterial colonies were picked form an LB/Agar-plate and either resuspended in PCR reaction mix and subsequently used to inoculate LB-medium with appropriate antibiotic, or resuspended in water. The water was then taken as "template" for PCR and inoculation of Minepreps.<br/> | ||
| + | The PCR was performed using 20 pmol of forward and reverse primer (= 0.2 µL of 100µM) in a total volume of 20 µL.<br/> | ||
| + | PCR conditions were set to recommendations made by Fermentas: | ||
| + | |||
| + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" | ||
| + | |||
| + | !Temp !! Time !! | ||
| + | |- | ||
| + | |95°||03:00|| | ||
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|94°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:30||rowspan="3"|x 30 cycles | ||
| + | |- | ||
| + | |45°'''*'''||style="border-right:solid 1px #000000;"|00:30 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|01:00 /kb | ||
| + | |- | ||
| + | |72°||10:00|| | ||
| + | |- | ||
| + | |4°||8|| | ||
| + | |} | ||
<br><br> | <br><br> | ||
| + | |||
| + | '''*'''with 45°C at a minimum of primer annealing temp | ||
| + | |||
| + | ===Plasmid-PCR=== | ||
| + | |||
| + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" | ||
| + | |||
| + | !Temp !! Time !! | ||
| + | |- | ||
| + | |98°||05:00|| | ||
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|98°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:10||rowspan="3"|x 35 cycles | ||
| + | |- | ||
| + | |60°||style="border-right:solid 1px #000000;"|00:30 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|02:30 | ||
| + | |- | ||
| + | |72°||10:00|| | ||
| + | |} | ||
| + | |||
| + | ===shRNA-PCR protocol=== | ||
| + | |||
| + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" | ||
| + | |||
| + | !Temp !! Time !! | ||
| + | |- | ||
| + | |98°||00:30|| | ||
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|98°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:10||rowspan="3"|x 35 cycles | ||
| + | |- | ||
| + | |60°||style="border-right:solid 1px #000000;"|00:30 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|00:15 | ||
| + | |- | ||
| + | |72°||1:00|| | ||
| + | |- | ||
| + | |4°||forever|| | ||
| + | |} | ||
===Gel extraction=== | ===Gel extraction=== | ||
After gel electrophoresis the digested vector and insert have to be purified from the gel. With the help of a UV lamp, the bands were quickly excised from the gel without exposing the DNA too long to UV light. Afterwards the DNA was purified with the QIAquick Gel extraction kit. Three volumes of buffer QG were added to one volume of gel. The gel fragment was dissolved by incubation for 10 min at 50°C. Afterwards one volume of 100% isopropanol was added. The solution was applied on a QIAquick spin column after this has been placed into a provided 2 ml collection tube. By centrifugation for 1 min at 13.000 rpm the DNA was bound to the column. The flow-through was discarded and the column was placed in the same collection tube. To remove all traces of agarose from the column, 500 µl of wash buffer QC was added followed by centrifugation for 1 min at 13.000 rpm. The flow-through was discarded and the column was washed with 750 µl of buffer PE for 1 min at 13.000 rpm. Afterwards the flow-through was discarded. An additional centrifugation for 1 min at 13.000 rpm helped to remove the residual ethanol. The column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 30 µl of ddH2O. | After gel electrophoresis the digested vector and insert have to be purified from the gel. With the help of a UV lamp, the bands were quickly excised from the gel without exposing the DNA too long to UV light. Afterwards the DNA was purified with the QIAquick Gel extraction kit. Three volumes of buffer QG were added to one volume of gel. The gel fragment was dissolved by incubation for 10 min at 50°C. Afterwards one volume of 100% isopropanol was added. The solution was applied on a QIAquick spin column after this has been placed into a provided 2 ml collection tube. By centrifugation for 1 min at 13.000 rpm the DNA was bound to the column. The flow-through was discarded and the column was placed in the same collection tube. To remove all traces of agarose from the column, 500 µl of wash buffer QC was added followed by centrifugation for 1 min at 13.000 rpm. The flow-through was discarded and the column was washed with 750 µl of buffer PE for 1 min at 13.000 rpm. Afterwards the flow-through was discarded. An additional centrifugation for 1 min at 13.000 rpm helped to remove the residual ethanol. The column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 30 µl of ddH2O. | ||
<br><br> | <br><br> | ||
| + | |||
| + | ===Large scale preparation of plasmid DNA=== | ||
| + | 150 ml LB-Medium with 150 µl ampicillin was inoculated with 50 µl of bacteria culture which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin MAxiprep kit from Qiagen and the protocol was followed. The overnight culture was centrifuged for 20 min at 4000 rpm at 4°C using an SLA 1500 Rotor. Afterwards the LB-medium was discarded and the pellet was homogeneously resuspended in 10 ml of precooled Buffer P1. After having added 10 ml of Buffer P2 the mixture was inverted 4-6 times and incubated for 5 min at RT before adding 10 ml of chilled Buffer P3. Thereafter the lysate was poured into a prepared QIAfilter Maxi Cartridge and incubated at RT for 10 min. During this time a QIAGEN-tip 500 was equilibrated by applying 10 ml of Buffer QBT and allowing the column to empty by gravity flow. The cell lysate was filtered into the QIAGEN-tip. The cleared lysate entered the resin by gravity flow and after washing with 2 x 30 ml Buffer QC the Plasmid DNA was eluted with 15 ml Buffer QF. After this the DNA was precipitated by adding 10.5 ml isopropanol and centrifuged at 4,000 rpm for 45 min at 4°C. The supernatant was discarded and the DNA pellet was washed with 5 ml ethanol (70%) and centrifuged at 4,000 rpm for 15 min. After air-drying the pellet the DNA was redissolved in H2O. <br> | ||
| + | <br> | ||
| + | |||
| + | ===Plasmid-DNA isolation=== | ||
| + | 5 ml LB-Medium with 5 µl ampicillin was inoculated with single colonies which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin Miniprep kit from Qiagen and following the manufacturer’s protocol. 4 ml of each overnight culture was pelleted in 2 ml microcentrifuge tubes during two steps of centrifugation at 13.000 rpm. Subsequently the pellet was resuspended in 250 µl of chilled buffer P1. 250 µl of lysis buffer P2 was added and the solution was mixed thoroughly by inverting the tube 4-6 times. After adding 350 µl of the neutralization Buffer N3 the solution was mixed immediately and thoroughly by inverting the tube 4-6 times. Thereafter the mixture was centrifuged. The supernatants were applied to a QIAprep column which was put in a 2 ml collection tube. It was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. After adding 500 µl of wash buffer PB, it was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. Once more, it was washed with 750 µl of wash buffer PE. In an additional centrifugation for 1 min at 13.000 rpm the residual wash buffer was removed. The QIAprep column was placed into a clean 1.5 ml microcentrifuge tube and the plasmid DNA was eluted in 30 µl ddH2O. | ||
| + | <br><br> | ||
| + | |||
| + | ===Preparation of competent E. coli Top10 and DH5alpha=== | ||
| + | Plating of E. coli Top10 and DH5alpha on a agar plate (LB, without Amp); Preperation of competent cells according to the following protocol: | ||
| + | |||
| + | First, a 20 ml over night culture was inoculated in antibiotic free LB medium from a fresh single colony and transferred into 400 ml antibiotic free LB medium the next day. This culture was incubated at 37 °C while shacking until an OD600 of 0.5 – 0.6 was achieved. The culture was than cooled down on ice, centrifuged (8 min, 4 °C, 3500 rpm), the supernatant discarded and the pellet resuspended in 10 ml 100 mM CaCl2. After addition of further 190 ml 100 mM CaCl2 the suspension was incubated on ice for 30 min. The suspension was than again centrifuged (8 min, 4 °C, 3500 rpm), the supernatant discarded, the pellet resuspended in 20 ml 82.5 mM CaCl2 with 17.5 % glycerol and aliquoted. The aliquots were flash frozen in liquid nitrogen and than stored at -80 °C until usage. | ||
| + | <br /><br /> | ||
===Purification of PCR product=== | ===Purification of PCR product=== | ||
| Line 21: | Line 99: | ||
<br><br> | <br><br> | ||
| - | === | + | |
| - | + | ===Sequencing=== | |
| + | Performed by the GATC Biotech company | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
<br><br> | <br><br> | ||
| - | === | + | == random assembly PCR (raPCR) == |
| - | + | ||
| - | <br> | + | ==== Designing Oligos ==== |
| + | |||
| + | To design Oligos for raPCR, first you need to have your miRNA sequence. This you can find on databases such as [http://www.microrna.org microRNA.org] or [http://mirbase.org mirbase.org]. Then you need to have an innert spacer sequence to seperate the binding sites from each other. | ||
| + | Like we did, you can use following tools to create your own spacer sequence: | ||
| + | :[http://cbit.snu.ac.kr/~miTarget/ http://cbit.snu.ac.kr/~miTarget/] | ||
| + | :[http://cbcsrv.watson.ibm.com/rna22.html http://cbcsrv.watson.ibm.com/rna22.html] | ||
| + | :[http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html] | ||
| + | |||
| + | Or you choose the one we used: <span style="color:#009be1">GCATACATGGACTGC</span><span style="color:#78b41e">CACTGAATCCAACTG</span> | ||
| + | |||
| + | Then the spacer need to be added to the miRNA binding site you want to use. In our approach, the spacer is also used for annealing of the different oligos. Therefore we suggest to split it in two parts. The first half (green) is then added to the 5' end and the second half (blue) is added to the 3' end of the miRBS sequence. | ||
| + | |||
| + | For running the PCR you need to create annealing oligos, which could either be the spacer itself (reverse complement of firsthalf elongated with reverse complement of second half). | ||
| + | |||
| + | Last but not least you need to have stop oligos. Stop oligos are characterised by having just one annealing sequence so, either first half or second half. Like for normal PCR Primers one annealing sequence should be reverse complemented, so they can also act effeciently as PCR primers. Additionally you can add whatever you want (i.e. the BBb-Standard sites). Those primers you can find in our [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#Primer_Table_raPCR primer list]. | ||
| + | |||
| + | |||
| + | ==== running the PCR ==== | ||
| + | |||
| + | <br \> | ||
| + | We developed a standardized PCR based method we use for the creation of our microRNA binding site patterns. It consists of two subsequent PCR steps: a 12 cycle PCR followed by a 25 cycle PCR. Therefor, order the oligos containing binding sites for the different microRNAs of your choice (refer to step 1). Furthermor, order the following diraPCR stop oligos: | ||
| + | :*miraPCR_Stop(fw)_EcoRI: 5'-TTTGTCgaattcGGTAGCTATTTCTC-3' | ||
| + | :*miraPCR_Stop(rev)_PstI: 5'-TTTGTCctgcagGAGAAATAGCTACC-3' | ||
| + | *Dilute all oligos (including the stop oligos) to a final concentration of 100 um. | ||
| + | *set up a PCR-reaction according to the following protocol. You can direct the length of your plasmid by applying more stop oligos (= shorter PCR fragments, that means patterns with lower amount of miR binding sites) and different amounts of binding site oligos. | ||
| + | <br \> | ||
| + | ''' 12-cycle PCR ''' | ||
| + | *set up a PCR reaction according to the following protocol | ||
| + | <br /> | ||
| + | :*0.1-4 µl of Oligos (recommended: for binding site patterns of the same miRNA, use 3 µL) | ||
| + | :*0-0.5 µLof each stop oligo (recommended: for shorter fragments use 0.5 µL) | ||
| + | :*1 µL of spacer oligo | ||
| + | :*25 µl of Phusion PCR MasterMix | ||
| + | :*add water to a final volume of 50 µl | ||
| + | <br /> | ||
| + | * run 12-cycle PCR according to the following program | ||
| + | |||
| + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" | ||
| + | |||
| + | !Temp !! Time !! | ||
| + | |- | ||
| + | |95°||05:00|| | ||
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|95°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:30||rowspan="3"|x 12 cycles | ||
| + | |- | ||
| + | |57°||style="border-right:solid 1px #000000;"|00:45 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|00:45 | ||
| + | |- | ||
| + | |4°||forever|| | ||
| + | |} | ||
| + | |||
| + | <br /> | ||
| + | *PCR purify all products applying a PCR purification Kit (Qiagen). We recommend elution in 30 µL for higher DNA concentration. | ||
| + | <br /> | ||
| + | '''25-cycle PCR''' | ||
| + | * run 25-cycle PCR in order to amplify constructs; therefor set up a PCR reaction according to the following protocol | ||
| + | <br /> | ||
| + | :*1-20 µl of purified PCR product (for shorter patterns (100-500 bp, 20 ul are recommended) | ||
| + | :*0.5-1 µl of each stop oligo | ||
| + | :*25 µl of Phusion PCR MasterMix (2x) | ||
| + | :*add water to final volume of 50 µl | ||
| + | <br /> | ||
| + | *run 25-cycle PCR according to the following protocol | ||
| + | <br /> | ||
| + | |||
| + | {| border="1" cellpadding="3" style="border:solid 1px #AAAAAA; border-collapse:collapse; background-color:#F9F9F9; empty-cells:show;" | ||
| + | |||
| + | !Temp !! Time !! | ||
| + | |- | ||
| + | |95°||05:00|| | ||
| + | |- | ||
| + | |style="border-top:solid 1px #000000;"|95°||style="border-top:solid 1px #000000;border-right:solid 1px #000000;"|00:30||rowspan="3"|x 25 cycles | ||
| + | |- | ||
| + | |65°||style="border-right:solid 1px #000000;"|00:45 | ||
| + | |- | ||
| + | |style="border-bottom:solid 1px #000000;"|72°||style="border-bottom:solid 1px #000000;border-right:solid 1px #000000;"|00:50 | ||
| + | |- | ||
| + | |72°||05:00|| | ||
| + | |- | ||
| + | |4°||forever|| | ||
| + | |} | ||
| + | <br><br> | ||
==Cell culture== | ==Cell culture== | ||
| Line 48: | Line 215: | ||
*take 1/10th of the cell suspension and plate out on the according dish (either p100 dish, 6 well plate or T-flask) | *take 1/10th of the cell suspension and plate out on the according dish (either p100 dish, 6 well plate or T-flask) | ||
<br> | <br> | ||
| + | |||
| + | ===Thawing=== | ||
| + | Thawing of Huh-7, HeLa p4 and HEK-293 cells according to the following protocol | ||
| + | vials from liquid nitrogen were thawn at 37 °C | ||
| + | once, the probe was nearly completely thawn, cells were thrown into pre-warmed DMEM (10 % FCS, L-Glut, P/S) and gentely mixed | ||
| + | cells were spinned down at 800 rpm, 3 min; supernatant was discarded | ||
| + | the pellett was resuspended in 10 ml DMEM an plated on a p100 cell culture dish in the following media according to the different cell lines: | ||
===Coating=== | ===Coating=== | ||
| - | + | For 96-well plates: | |
*add 30 µl of poly-L-lysine solution (Sigma) to each well (make sure whole surface is covered with solution) | *add 30 µl of poly-L-lysine solution (Sigma) to each well (make sure whole surface is covered with solution) | ||
*leave for 30' in the incubator | *leave for 30' in the incubator | ||
| Line 111: | Line 285: | ||
*add transfection mix to media (1 volume of mix, 2 volumes of media) | *add transfection mix to media (1 volume of mix, 2 volumes of media) | ||
*pour on cells | *pour on cells | ||
| + | <br> | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
<br> | <br> | ||
| Line 138: | Line 318: | ||
To find out the concentration of viral genomes per ml (<font face="Symbol">Y</font>) the value of the <font face="Symbol">X</font> position on the standard curve has to be multiplied as follows: <font face="Symbol">Y</font> = <font face="Symbol">X</font> ×10² ×10² | To find out the concentration of viral genomes per ml (<font face="Symbol">Y</font>) the value of the <font face="Symbol">X</font> position on the standard curve has to be multiplied as follows: <font face="Symbol">Y</font> = <font face="Symbol">X</font> ×10² ×10² | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| + | <br> | ||
== Measurements == | == Measurements == | ||
=== ELISA === | === ELISA === | ||
| Line 185: | Line 371: | ||
=== Microscopy === | === Microscopy === | ||
| - | + | We used fluorescence microscopy to measure EGFP and EBFP2 fluorescence intensity. Transfection efficiency was first evaluated using epifluorescence microscopes (Leica DM IRB or Olympus IX81) directly on the 96-well plates. Only cells which were transfected successfully were measured. Cells were washed with 1x PBS and detached from the plate using trypsin. 30µl trypsin was added to each well. After incubation for ten minutes at room temperature, ells were resuspended in 170µl 1%BSA in PBS and replicates for each condition were pooled into 8-well coverslip chambers. 100-150µl were used for confocal microscopy. | |
| - | Single images were obtained using | + | Single cell images were obtained using a Leica TCS SP5 laser scanning confocal microscope (LSCM) and alternatively a Leica TCS SP2 LSCM. EGFP fluorescence was excited by the 488nm laserline of an Argon laser and measured between 520 and 560nm, EBFP2 proteins were excited by UV laser at 405nm and measured between 440 and 460nm. Pictures were taken sequentially line by line in two different channels for EGFP, EBFP2. Bright field was acquired at the same time as the EBFP2 signal from the 405nm laser. |
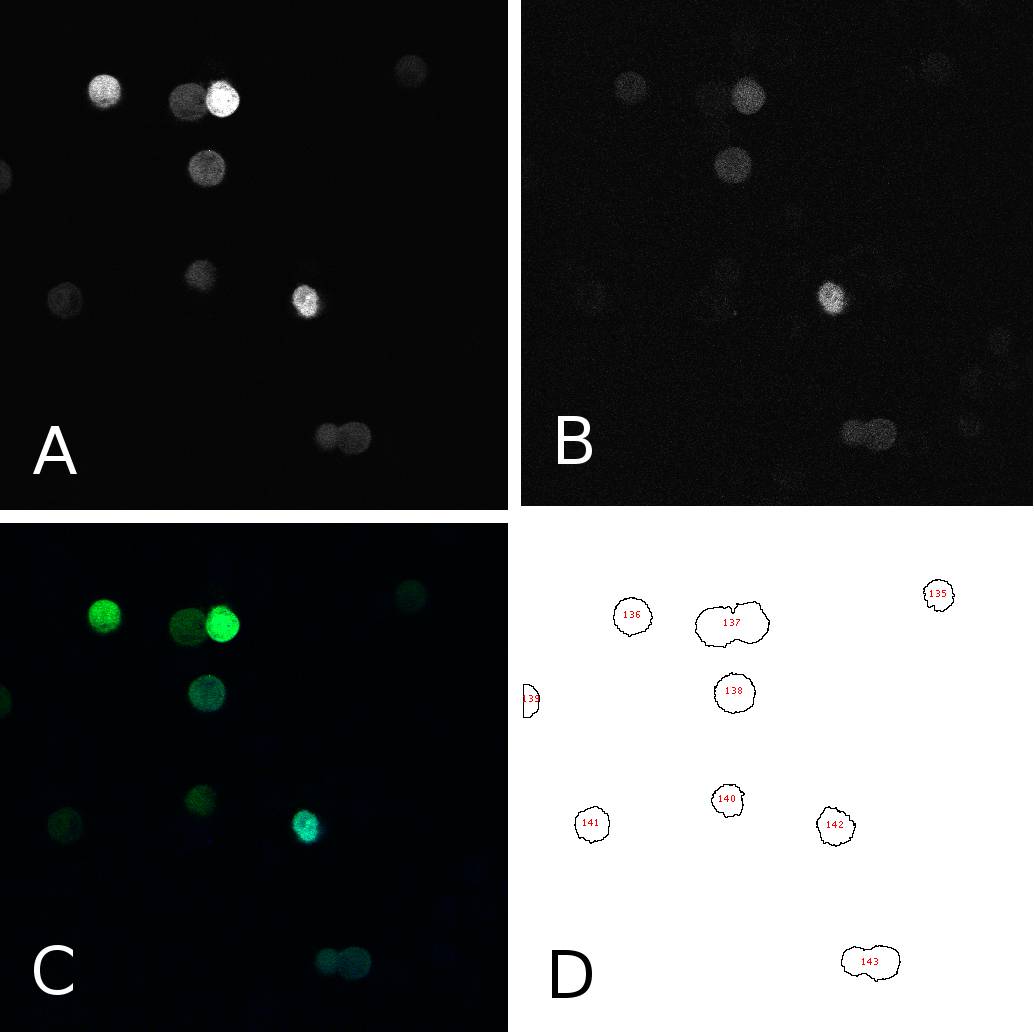
[[Image:panel.jpg|thumb|400px|center|'''HeLa cells two days after transfection with miMeasure''' (A) fluorescence signal GFP channel, 8bit; (B) fluorescence signal BFP channel, 8bit; (C) merge of channels A and B, RGB (D) cells after segmentation and automated cell counting and annotation]] | [[Image:panel.jpg|thumb|400px|center|'''HeLa cells two days after transfection with miMeasure''' (A) fluorescence signal GFP channel, 8bit; (B) fluorescence signal BFP channel, 8bit; (C) merge of channels A and B, RGB (D) cells after segmentation and automated cell counting and annotation]] | ||
| - | To analyze the fluorescence of single cells | + | To analyze the blue and green fluorescence of single cells we analyzed the images with ''ImageJ''. First, we separated the intensities coming from the bright cells from the background by thresholding the images. This allowed us to annotate cells automatically using the “analyze particles” tool. Measurements were redirected to the EGFP or EBFP2 respectively and the mean intensities were for each cell in each channel were saved. Panel 1 shows an example of one such image in different channels and after segmentation. From the data thus obtained, we calculated the EGFP:EBFP2 ratios for each cell using a simple algorithm. The average of these ratios served as the comparison measure and was visualized in bar plots for different constructs and/or conditions. Alternatively we used to individual data points to calculate the correlation between EGFP and EBFP2 expression through a simple linear regression analysis. |
* Consumables and Chemicals | * Consumables and Chemicals | ||
PerkinElmer ViewPlate, product number: 6004920<br> | PerkinElmer ViewPlate, product number: 6004920<br> | ||
| - | Nunc | + | Nunc Lab-Tek coverglass chamber, product number: 155411<br> |
1x PBS (1.37mM NaCl, 0.27mM KCl, 10mM Na2HPO4, 0.2mM KH2PO4)<br> | 1x PBS (1.37mM NaCl, 0.27mM KCl, 10mM Na2HPO4, 0.2mM KH2PO4)<br> | ||
0.05% Trypsin-EDTA, Invitrogen GIBCO, product number: 15400054<br> | 0.05% Trypsin-EDTA, Invitrogen GIBCO, product number: 15400054<br> | ||
| Line 203: | Line 389: | ||
Leica DM IRB<br> | Leica DM IRB<br> | ||
Leica SP5<br> | Leica SP5<br> | ||
| + | Leica SP2<br> | ||
ImageJ version 1.43<br> | ImageJ version 1.43<br> | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
<br> | <br> | ||
| + | |||
| + | === Flow cytometry === | ||
| + | |||
| + | We used flow cytometry to analyse the EGFP and EBFP2 fluorescence intensities. Cells grown in a 96 well plate were washed once with 1xPBS and then trypsinised. Afterwards they were resuspended in PBS + 1% BSA and analyzed. EGFP and EBFP2 cellular intensities were measured with a flow cytometer was performed equipped with two laser diodes, one at 405nm and one at 488nm. The fluorescence of EGFP and EBFP2 was separated at 495nm by a dichroic mirror. Each signal was filtered through two emission filters: the first with the band pass between 425nm and 475nm to detect EBFP2, the second between 497.5nm and 522.5nm to detect EGFP. The bleed through was compensated by measuring cells only expressing EGFP and accordingly EBFP2. For the measurements 100µl cells in PBS + 1% BSA were pumped by the machine and about 10000 cells were analysed for each construct. | ||
| + | |||
| + | |||
| + | * Consumables and Chemicals | ||
| + | Falcon Becton Dickinson Microtest 96, product number: 353072 <br> | ||
| + | 1x PBS (1.37mM NaCl, 0.27mM KCl, 10mM Na2HPO4, 0.2mM KH2PO4) <br> | ||
| + | Bovine Serum Albumine, Sigma, product number: A9647 <br> | ||
| + | |||
| + | |||
| + | * Instruments | ||
| + | Beckman Coulter Cytomics FC500MPL | ||
| + | |||
| + | |||
| + | *Analysis | ||
| + | The reduction of EGFP expression compared to EBFP2 that we observed by flow cytometry could be quantitated as follow: two-dimensional scatter count plots representing cell counts for each EBFP2-EGFP intensity pair on a linear scale were exported as 64X64 pixel images. To ignore non-transfected cells, a square of 3X3 pixel originating from (0,0), where control non-transfected cells accumulate, was put to zero in each count plot. A ratio image of the same size containing the EGFP-EBFP2 intensity ratio for each pixel was generated. The mean ratio was estimated by multiplying each count plot with the ratio image, measuring the total image intensity and dividing it by the total count of each count plot. For the standard deviation, the mean intensity of a count plot was subtracted to the ratio image. the square of each pixel intensity was multiplied by the corresponding count and the total intensity of the result was divided by the total count. The standard deviation was then calculated by taking the square root of this number. All steps were performed on imageJ. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
=== Dual Luciferase Assay === | === Dual Luciferase Assay === | ||
We measured the knockdown of firefly luciferase using the [http://www.promega.com/tbs/tm046/tm046.pdf Promega Dual Luciferase Reporter Assay]. | We measured the knockdown of firefly luciferase using the [http://www.promega.com/tbs/tm046/tm046.pdf Promega Dual Luciferase Reporter Assay]. | ||
| Line 216: | Line 436: | ||
The activity of the first luciferase (firefly) was measured by adding 25µl of LAR II reagent to the well. The enzyme reacts upon translation without further processing and oxidates beetle luciferin, resulting in photon emission that can be measured. In addition to beetle luciferin, the LAR II reagent contains coenzyme A, which accelerates the reaction and thus creates a prolonged luminescence signal. The luminescence was measured two seconds after addition of the reagent, for ten seconds. Afterwards, 25µl Stop & Glo reagent was added, which is able to quench the firefly luciferase activity and simultaneously contains the substrate for Renilla luciferase, coelenterazine. This second reaction also emits photons upon oxidation of the substrate. Addition of substrates and light emission measurements were conducted automatically by the GLOMAX Luminometer. | The activity of the first luciferase (firefly) was measured by adding 25µl of LAR II reagent to the well. The enzyme reacts upon translation without further processing and oxidates beetle luciferin, resulting in photon emission that can be measured. In addition to beetle luciferin, the LAR II reagent contains coenzyme A, which accelerates the reaction and thus creates a prolonged luminescence signal. The luminescence was measured two seconds after addition of the reagent, for ten seconds. Afterwards, 25µl Stop & Glo reagent was added, which is able to quench the firefly luciferase activity and simultaneously contains the substrate for Renilla luciferase, coelenterazine. This second reaction also emits photons upon oxidation of the substrate. Addition of substrates and light emission measurements were conducted automatically by the GLOMAX Luminometer. | ||
<br> | <br> | ||
| + | ==== Order of recording and processing raw data ==== | ||
| + | # triple-transfection of the following constructs in ratio 10:1:1 | ||
| + | #* shRNA-like miRNA (shRNA miR) expressing plasmid (25 ng / well) | ||
| + | #* reference construct containing hRluc (2.5 ng / well) | ||
| + | #* tuning construct with Luc2 fused to binding site referring to shRNA miR (2.5 ng / well) | ||
| + | #: note: used backbones for all constructs were either pBS_U6 or pBS_H1 | ||
| + | # measurement of bioluminescent signal after substrate addition | ||
| + | #: raw data file with following <html><a name="nomenclature" href="#nomenclature">nomenclature</a></html>: shRNA_date_samples_ctrl_BB.xls | ||
| + | ## first injection: LARII | ||
| + | ##* quantification of Firefly signal | ||
| + | ## second injection: Stop & Glo® Reagent | ||
| + | ##* quantification of Renilla signal | ||
| + | # calculate signal ratio: Firefly/Renilla | ||
| + | #: note: Renilla is used as a reference, Firefly signal determines binding site strength | ||
| + | #* controls on every plate: perfect binding site (lower expression limit), no binding site (upper expression limit = 100%) defining overall expression range | ||
| + | #* another control: separate triple-transfections of tuning constructs with a shRNA miR expression plasmid that is ''not'' corresponding to the binding site on the tuning construct (upper expression limit) | ||
| + | # normalization by control without binding site | ||
| + | #* thus, normalized value corresponds to knockdown percentage | ||
| + | # deviation of knockdown percentage for corresponding shRNA miR by referring knockdown percentage of non-corresponding shRNA miR | ||
| + | # calculate mean values and standard deviation (out of eight replicates) for each construct containing imperfect binding site (intermediate expression) | ||
| + | #: raw data file with following <html><a name="nomenclature" href="#nomenclature">nomenclature</a></html>: plate#_process_BB_shRNA.xls | ||
| + | #* note: error propagation has to be taken into account | ||
| + | # grade calculated knockdown percentages in ascending order (lowest value always for perfect binding site, whereas maximum value without binding site) | ||
| + | #: raw data file with following <html><a name="nomenclature" href="#nomenclature">nomenclature</a></html>: shRNA_BB_final.xlsx | ||
| + | <br/> | ||
| + | ===== nomenclature ===== | ||
| + | {| class="wikitable sortable" border="0" style="text-align: left" | ||
| + | |-bgcolor=#cccccc | ||
| + | |+ align="top, left"|'''table 1''': Used nomenclature for raw data files. | ||
| + | | abbreviation||meaning||example | ||
| + | |- | ||
| + | |shRNA||expressed shRNA miR||shRNA miR against hAAT ("hAAT") | ||
| + | |- | ||
| + | |date||date of measurement in format YYYYMMDD||20101026 | ||
| + | |- | ||
| + | |samples||internal ID of constructs||M1-M6 | ||
| + | |- | ||
| + | |BB||plasmid backbone||either pBS_U6 ("U6") or pBS_H1 ("H1") | ||
| + | |- | ||
| + | |plate#||number of measured 96 well plate||plate1 | ||
| + | |- | ||
| + | |} | ||
| + | <br/> | ||
==== Consumables and Reagents ==== | ==== Consumables and Reagents ==== | ||
| Line 224: | Line 487: | ||
==== Instruments ==== | ==== Instruments ==== | ||
Promega GLOMAX 96 Microplate Luminometer | Promega GLOMAX 96 Microplate Luminometer | ||
| - | |||
| - | == | + | <html> |
| - | + | <div class="backtop"> | |
| - | + | <a href="#top">↑</a> | |
| - | + | </div> | |
| - | + | </html> | |
| - | + | <br><br> | |
| - | + | {{:Team:Heidelberg/Single_Bottom}} | |
| - | + | ||
| - | {{:Team:Heidelberg/ | + | |
Latest revision as of 02:32, 28 October 2010

MethodsCloningAgarose Gel ElectrophoresisAgarose flat-bed gels in various concentrations (0.6–2% agarose in 0.5 x TAE buffer) and sizes were run to separate DNA fragments in an electrical field (10–20 V/cm) for analytical or preparative use. The desired amount of agarose was boiled in 1 x TAE buffer until it was completely dissolved. After it cooled down to approximately 60°C, ethidium bromide (EtBr) solution (0.5 µg/ml final concentration) was added to the liquid agar, which was then poured in a flat-bed tray with combs. As soon as the agarose solidified, the Running buffer (0.5 x TAE buffer) was added before the DNA in the loading buffer was loaded into the wells and separated electrophoretically. Ethidium bromide intercalates with the DNA’s GC ntss resulting in DNA-EtBr-complex that emits visible light. Therefore, the DNA fragments could be detected on a UV-light tray at 265 nm.
Colony PCRColony PCRs were performed using Fermentas PCR Master Mix (2x), containing Taq DNA Polymerase.
*with 45°C at a minimum of primer annealing temp Plasmid-PCR
shRNA-PCR protocol
Gel extractionAfter gel electrophoresis the digested vector and insert have to be purified from the gel. With the help of a UV lamp, the bands were quickly excised from the gel without exposing the DNA too long to UV light. Afterwards the DNA was purified with the QIAquick Gel extraction kit. Three volumes of buffer QG were added to one volume of gel. The gel fragment was dissolved by incubation for 10 min at 50°C. Afterwards one volume of 100% isopropanol was added. The solution was applied on a QIAquick spin column after this has been placed into a provided 2 ml collection tube. By centrifugation for 1 min at 13.000 rpm the DNA was bound to the column. The flow-through was discarded and the column was placed in the same collection tube. To remove all traces of agarose from the column, 500 µl of wash buffer QC was added followed by centrifugation for 1 min at 13.000 rpm. The flow-through was discarded and the column was washed with 750 µl of buffer PE for 1 min at 13.000 rpm. Afterwards the flow-through was discarded. An additional centrifugation for 1 min at 13.000 rpm helped to remove the residual ethanol. The column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 30 µl of ddH2O.
Large scale preparation of plasmid DNA150 ml LB-Medium with 150 µl ampicillin was inoculated with 50 µl of bacteria culture which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin MAxiprep kit from Qiagen and the protocol was followed. The overnight culture was centrifuged for 20 min at 4000 rpm at 4°C using an SLA 1500 Rotor. Afterwards the LB-medium was discarded and the pellet was homogeneously resuspended in 10 ml of precooled Buffer P1. After having added 10 ml of Buffer P2 the mixture was inverted 4-6 times and incubated for 5 min at RT before adding 10 ml of chilled Buffer P3. Thereafter the lysate was poured into a prepared QIAfilter Maxi Cartridge and incubated at RT for 10 min. During this time a QIAGEN-tip 500 was equilibrated by applying 10 ml of Buffer QBT and allowing the column to empty by gravity flow. The cell lysate was filtered into the QIAGEN-tip. The cleared lysate entered the resin by gravity flow and after washing with 2 x 30 ml Buffer QC the Plasmid DNA was eluted with 15 ml Buffer QF. After this the DNA was precipitated by adding 10.5 ml isopropanol and centrifuged at 4,000 rpm for 45 min at 4°C. The supernatant was discarded and the DNA pellet was washed with 5 ml ethanol (70%) and centrifuged at 4,000 rpm for 15 min. After air-drying the pellet the DNA was redissolved in H2O. Plasmid-DNA isolation5 ml LB-Medium with 5 µl ampicillin was inoculated with single colonies which grew overnight on a shaker at 37°C. The plasmid DNA was isolated using QiAprep Spin Miniprep kit from Qiagen and following the manufacturer’s protocol. 4 ml of each overnight culture was pelleted in 2 ml microcentrifuge tubes during two steps of centrifugation at 13.000 rpm. Subsequently the pellet was resuspended in 250 µl of chilled buffer P1. 250 µl of lysis buffer P2 was added and the solution was mixed thoroughly by inverting the tube 4-6 times. After adding 350 µl of the neutralization Buffer N3 the solution was mixed immediately and thoroughly by inverting the tube 4-6 times. Thereafter the mixture was centrifuged. The supernatants were applied to a QIAprep column which was put in a 2 ml collection tube. It was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. After adding 500 µl of wash buffer PB, it was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded. Once more, it was washed with 750 µl of wash buffer PE. In an additional centrifugation for 1 min at 13.000 rpm the residual wash buffer was removed. The QIAprep column was placed into a clean 1.5 ml microcentrifuge tube and the plasmid DNA was eluted in 30 µl ddH2O.
Preparation of competent E. coli Top10 and DH5alphaPlating of E. coli Top10 and DH5alpha on a agar plate (LB, without Amp); Preperation of competent cells according to the following protocol: First, a 20 ml over night culture was inoculated in antibiotic free LB medium from a fresh single colony and transferred into 400 ml antibiotic free LB medium the next day. This culture was incubated at 37 °C while shacking until an OD600 of 0.5 – 0.6 was achieved. The culture was than cooled down on ice, centrifuged (8 min, 4 °C, 3500 rpm), the supernatant discarded and the pellet resuspended in 10 ml 100 mM CaCl2. After addition of further 190 ml 100 mM CaCl2 the suspension was incubated on ice for 30 min. The suspension was than again centrifuged (8 min, 4 °C, 3500 rpm), the supernatant discarded, the pellet resuspended in 20 ml 82.5 mM CaCl2 with 17.5 % glycerol and aliquoted. The aliquots were flash frozen in liquid nitrogen and than stored at -80 °C until usage.
Purification of PCR productOne volume of buffer PBI was added to one volume of the PCR sample mix. The sample was applied to a QIAquick column which has been placed into a provided 2 ml collection tube. It was centrifuged for 1 min at 13.000 rpm and the flow-through was discarded and the column was placed in the same collection tube. After this 750 µl of buffer PE was added to wash the column. It was centrifuged for 1 min at 13.000 rpm. The flow-through was discarded and the column was placed in the same collection tube. It was centrifuged for 1 min at 13.000 rpm. Afterwards the QIAquick column was placed into a new 1.5 ml microcentrifuge tube and it was eluted with 40 µl ddH2O.
SequencingPerformed by the GATC Biotech company
random assembly PCR (raPCR)Designing OligosTo design Oligos for raPCR, first you need to have your miRNA sequence. This you can find on databases such as [http://www.microrna.org microRNA.org] or [http://mirbase.org mirbase.org]. Then you need to have an innert spacer sequence to seperate the binding sites from each other. Like we did, you can use following tools to create your own spacer sequence:
Or you choose the one we used: GCATACATGGACTGCCACTGAATCCAACTG Then the spacer need to be added to the miRNA binding site you want to use. In our approach, the spacer is also used for annealing of the different oligos. Therefore we suggest to split it in two parts. The first half (green) is then added to the 5' end and the second half (blue) is added to the 3' end of the miRBS sequence. For running the PCR you need to create annealing oligos, which could either be the spacer itself (reverse complement of firsthalf elongated with reverse complement of second half). Last but not least you need to have stop oligos. Stop oligos are characterised by having just one annealing sequence so, either first half or second half. Like for normal PCR Primers one annealing sequence should be reverse complemented, so they can also act effeciently as PCR primers. Additionally you can add whatever you want (i.e. the BBb-Standard sites). Those primers you can find in our primer list.
running the PCR
Cell cultureMedia
Passaging
ThawingThawing of Huh-7, HeLa p4 and HEK-293 cells according to the following protocol vials from liquid nitrogen were thawn at 37 °C once, the probe was nearly completely thawn, cells were thrown into pre-warmed DMEM (10 % FCS, L-Glut, P/S) and gentely mixed cells were spinned down at 800 rpm, 3 min; supernatant was discarded the pellett was resuspended in 10 ml DMEM an plated on a p100 cell culture dish in the following media according to the different cell lines: CoatingFor 96-well plates:
TransfectionFuGENEDAY1
DAY2
PEI150cm2 plate format
HBSS150cm2 flask format
Virus Production
Virus production was done in 150cm2 flasks. 0.9 million cells were seeded in 30ml medium per flask. Transfections were done as described above.
To harvest the cells, cell suspension was decanted into 500ml corning conical centrifuge tubes. Remaining cells were washed with 19ml PBS and also transferred into the tubes. Cells were collected by centrifugation at 1500rpm for 15min at 4°C. Supernatant was aspirated, cells resuspended in 10ml 1xPBS and transferred into a 50ml blue-cap vial. Centrifugaion as above, aspiration of supernatant. Pellet was then resuspended in 5ml virus lysate solution and frozen at -196°C in liquid nitrogen for 5min, then thawed at 37°C. This freeze/thaw cycle was repeated five times.
Each sample was sonicated in a sonication bath for 1min 20s, 50µg/ml benzonase was added. Samples were kept at 37°C for 30min and vortexed every 10min. Centrifugation for 15min at 3270g, 4°C. Supernatant was transferred in to a new blue cap vial with a 5ml pipette. Gradiant was poured in a Beckman Quick-Seal centrifuge tube and a pasteur pipette plugged into the tube. 5ml of the virus suspension was transferred through the Pasteur pipette into the tube. 1.5ml 15% Iodixanolsolution (in PBS-MK) was poured through the Pasteur pipette in a way thet it presses put the virus suspension. In the same manner, 1.5ml 25% Iodixanolsolution was pipette to become the lower phase, then 1.5ml 40% Iodixanolsolution and finally 1.5ml 60% Iodixanolsolution. The Pasteur pipette was reomeved and the tubes sealed with tube sealer. Ultracentrifulation for 2h at 50.000rpm, 4°C and 71,1 Ti. The virus was recovered from the 40% Iodixanol phase. Seeding medium: DMEM high glucose, 10% FCS, 1% P/S, 1% L-Glutamin Virus lysate solution PBS-MK: 1xPBS, 1mM MgCl2, 2.5mM KCl Iodixanolsolution
Quantitative Realtime PCRTo quantitavely measure the amount of AAV rep gene, viruses had to be lysed. First, 10µl TE buffer was mixed with 10µl AAV solution. 20µl 2M NaOH were added and incubated for 30min at 56°C to lyse the viruses. Then 38µl 1M HCl and 922µl H2O were added. For a negative control, the same procedure was done with 10µl H2O instead of virus. For each probe, a 35µl mix of RNase free water, 1x SensiMix II Probe PCR Master Mix, 100pmol/µl forward and reverse primer and 100pm/µl probe was prepared in RNase free water. This was enough for measurement triplicates. To make a RT-PCR standard curve, the standard probes were diluted to concentrations of 3.5x1011 to 3.5x103 molecules in 10µl. Thermocycler program was 10min at 95°C initial activation, followed by 40 cycles of 10s at 95°C and 20s at 60°C. To find out the concentration of viral genomes per ml (Y) the value of the X position on the standard curve has to be multiplied as follows: Y = X ×10² ×10²
MeasurementsELISAProcedure
Buffers
MicroscopyWe used fluorescence microscopy to measure EGFP and EBFP2 fluorescence intensity. Transfection efficiency was first evaluated using epifluorescence microscopes (Leica DM IRB or Olympus IX81) directly on the 96-well plates. Only cells which were transfected successfully were measured. Cells were washed with 1x PBS and detached from the plate using trypsin. 30µl trypsin was added to each well. After incubation for ten minutes at room temperature, ells were resuspended in 170µl 1%BSA in PBS and replicates for each condition were pooled into 8-well coverslip chambers. 100-150µl were used for confocal microscopy. Single cell images were obtained using a Leica TCS SP5 laser scanning confocal microscope (LSCM) and alternatively a Leica TCS SP2 LSCM. EGFP fluorescence was excited by the 488nm laserline of an Argon laser and measured between 520 and 560nm, EBFP2 proteins were excited by UV laser at 405nm and measured between 440 and 460nm. Pictures were taken sequentially line by line in two different channels for EGFP, EBFP2. Bright field was acquired at the same time as the EBFP2 signal from the 405nm laser.
To analyze the blue and green fluorescence of single cells we analyzed the images with ImageJ. First, we separated the intensities coming from the bright cells from the background by thresholding the images. This allowed us to annotate cells automatically using the “analyze particles” tool. Measurements were redirected to the EGFP or EBFP2 respectively and the mean intensities were for each cell in each channel were saved. Panel 1 shows an example of one such image in different channels and after segmentation. From the data thus obtained, we calculated the EGFP:EBFP2 ratios for each cell using a simple algorithm. The average of these ratios served as the comparison measure and was visualized in bar plots for different constructs and/or conditions. Alternatively we used to individual data points to calculate the correlation between EGFP and EBFP2 expression through a simple linear regression analysis.
PerkinElmer ViewPlate, product number: 6004920
Leica DM IRB
Flow cytometryWe used flow cytometry to analyse the EGFP and EBFP2 fluorescence intensities. Cells grown in a 96 well plate were washed once with 1xPBS and then trypsinised. Afterwards they were resuspended in PBS + 1% BSA and analyzed. EGFP and EBFP2 cellular intensities were measured with a flow cytometer was performed equipped with two laser diodes, one at 405nm and one at 488nm. The fluorescence of EGFP and EBFP2 was separated at 495nm by a dichroic mirror. Each signal was filtered through two emission filters: the first with the band pass between 425nm and 475nm to detect EBFP2, the second between 497.5nm and 522.5nm to detect EGFP. The bleed through was compensated by measuring cells only expressing EGFP and accordingly EBFP2. For the measurements 100µl cells in PBS + 1% BSA were pumped by the machine and about 10000 cells were analysed for each construct.
Falcon Becton Dickinson Microtest 96, product number: 353072
Beckman Coulter Cytomics FC500MPL
The reduction of EGFP expression compared to EBFP2 that we observed by flow cytometry could be quantitated as follow: two-dimensional scatter count plots representing cell counts for each EBFP2-EGFP intensity pair on a linear scale were exported as 64X64 pixel images. To ignore non-transfected cells, a square of 3X3 pixel originating from (0,0), where control non-transfected cells accumulate, was put to zero in each count plot. A ratio image of the same size containing the EGFP-EBFP2 intensity ratio for each pixel was generated. The mean ratio was estimated by multiplying each count plot with the ratio image, measuring the total image intensity and dividing it by the total count of each count plot. For the standard deviation, the mean intensity of a count plot was subtracted to the ratio image. the square of each pixel intensity was multiplied by the corresponding count and the total intensity of the result was divided by the total count. The standard deviation was then calculated by taking the square root of this number. All steps were performed on imageJ.
Dual Luciferase AssayWe measured the knockdown of firefly luciferase using the [http://www.promega.com/tbs/tm046/tm046.pdf Promega Dual Luciferase Reporter Assay].
The DLR™ Assay System provides an efficient mean of performing dual-reporter assays, where the activities of firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferases (RL) are measured sequentially from a single sample. Firefly and Renilla luciferases can be used as a good reporter system, as those two enzymes have dissimilar enzyme structures and substrate requirements. This allows for selective discrimination between their bioluminescent reactions. The firefly luciferase (FL) reporter is measured first by adding Luciferase Assay Reagent II (LAR II) to generate a stabilized luminescent signal. After quantifying the firefly luminescence, this reaction is quenched, and the Renilla luciferase reaction is simultaneously initiated by adding Stop & Glo® Reagent to the same tube. The Stop & Glo® Reagent also produces a stabilized signal from the Renilla luciferase, which decays slowly over the course of the measurement. Here, Renilla luciferase is used for normalization. The measurements were conducted on the Promega GLOMAX 96 Microplate Luminometer using the Promega standard protocol (Sherf et al., 1996). LAR II reagent was prepared by resuspending Luciferase Assay Substrate in 10ml Luciferase Assay Buffer II. For Stop & Glo reagent, 2.1ml 50x Stop & Glo substrate and 105ml Stop & Glo Buffer were added to the amber Stop & Glo reagent bottle and mixed by vortexing. Reagents where stored in 15ml aliquots at -80°C and thawed freshly prior to each measurement. To set up the Luminometer, the two injectors where flushed with distilled water, 70% ethanol, again water and air, three times each. Afterwards, they were primed three times with substrate reagents. The activity of the first luciferase (firefly) was measured by adding 25µl of LAR II reagent to the well. The enzyme reacts upon translation without further processing and oxidates beetle luciferin, resulting in photon emission that can be measured. In addition to beetle luciferin, the LAR II reagent contains coenzyme A, which accelerates the reaction and thus creates a prolonged luminescence signal. The luminescence was measured two seconds after addition of the reagent, for ten seconds. Afterwards, 25µl Stop & Glo reagent was added, which is able to quench the firefly luciferase activity and simultaneously contains the substrate for Renilla luciferase, coelenterazine. This second reaction also emits photons upon oxidation of the substrate. Addition of substrates and light emission measurements were conducted automatically by the GLOMAX Luminometer.
Order of recording and processing raw data
nomenclature
Consumables and ReagentsLumaPlate, PerkinElmer, catalogue number 6005630 InstrumentsPromega GLOMAX 96 Microplate Luminometer
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"