Team:Minnesota/Project
From 2010.igem.org
(→Results) |
|||
| (16 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
!align="center"|[[Team:Minnesota/Notebook|<font color="gold">Notebook</font>]] | !align="center"|[[Team:Minnesota/Notebook|<font color="gold">Notebook</font>]] | ||
!align="center"|[[Team:Minnesota/Judging|<font color="gold">Judging Criteria</font>]] | !align="center"|[[Team:Minnesota/Judging|<font color="gold">Judging Criteria</font>]] | ||
| + | !align="center"|[[Team:Minnesota/Attributions|<font color="gold">Attributions</font>]] | ||
!align="center"|[[Team:Minnesota/Safety|<font color="gold">Safety</font>]] | !align="center"|[[Team:Minnesota/Safety|<font color="gold">Safety</font>]] | ||
|} | |} | ||
| Line 15: | Line 16: | ||
== Introduction == | == Introduction == | ||
| - | [[Image:BMC Catalysis.gif|frame| | + | [[Image:BMC Catalysis.gif|frame|Figure 1. Proposed nanobioreactor model. (R = reactant, I = intermediate, P = product and E1, E2 = enzymes)]] |
A bacterial microcompartment (BMC) is a polyhedral protein complex that encapsulates enzymes involved in specific metabolic pathways. BMCs were first observed by microscope in cyanobacteria the 1950’s. By the late 1990’s, it had become clear that these proteinaceous structures are produced by many types of bacteria, and serve various functions (Yeates et al. 2008). There are multiple examples of BMC’s occurring in nature: the carboxysome found in cyanobacteria and chemoautotrophic bacteria, the propanediol utilization compartment found in enterobacteria, as well as the ethanolamine utilization (Eut) compartment used in this project, also found in enterobacteria. By sequestering enzymes and substrates together in an enclosed space within the cell, the overall reaction efficiency is greatly improved. In some species, BMCs localize toxic compounds with enzymes that convert them into benign compounds. In this way a bacterial cell can ensure that the problematic intermediate is not released into the cell cytoplasm. | A bacterial microcompartment (BMC) is a polyhedral protein complex that encapsulates enzymes involved in specific metabolic pathways. BMCs were first observed by microscope in cyanobacteria the 1950’s. By the late 1990’s, it had become clear that these proteinaceous structures are produced by many types of bacteria, and serve various functions (Yeates et al. 2008). There are multiple examples of BMC’s occurring in nature: the carboxysome found in cyanobacteria and chemoautotrophic bacteria, the propanediol utilization compartment found in enterobacteria, as well as the ethanolamine utilization (Eut) compartment used in this project, also found in enterobacteria. By sequestering enzymes and substrates together in an enclosed space within the cell, the overall reaction efficiency is greatly improved. In some species, BMCs localize toxic compounds with enzymes that convert them into benign compounds. In this way a bacterial cell can ensure that the problematic intermediate is not released into the cell cytoplasm. | ||
| - | BMCs are ideal for iGEM because they could be harnessed as nanobioreactors for metabolic engineering. Interest in BMCs and their natural functions has | + | BMCs are ideal for iGEM because they could be harnessed as nanobioreactors for metabolic engineering. Interest in BMCs and their natural functions has grown in recent years. With increasing availability of information of their properties, BMCs are becoming more practical for applications in synthetic biology. Engineering a BMC to house an enzymatic pathway would increase the efficiency of the process, and would also quarantine any toxic intermediates (Figure 1). |
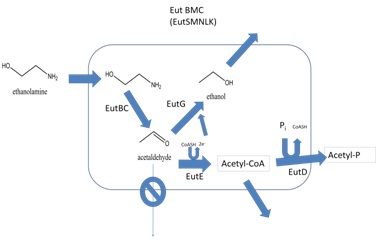
| - | Our goals were to develop E. coli that express a recombinant BMC, and develop a system for targeting proteins to be encapsulated within that compartment. We chose to use the Ethanolamine utilization (Eut) shell from Salmonella enterica because this species has been shown to produce these structures and they have been well characterized (Penrod & Roth, 2006). The ethanolamine utilization operon from Salmonella enterica strain LT2 is composed of 17 genes ( | + | Our goals were to develop E. coli that express a recombinant BMC, and develop a system for targeting proteins to be encapsulated within that compartment. We chose to use the Ethanolamine utilization (Eut) shell from Salmonella enterica because this species has been shown to produce these structures and they have been well characterized (Penrod & Roth, 2006). The ethanolamine utilization operon from Salmonella enterica strain LT2 is composed of 17 genes (Figure 2). Of these, only five possess BMC associated domains and are believed to be components of the Eut microcompartment (Figure 3). In Salmonella enterica this organelle-like structure localizes the enzymes required for ethanolamine utilization and prevents acetaldehyde, a volatile intermediate, from diffusing into the cytoplasm (Figure 4). |
{| | {| | ||
| - | [[Image:Eut Operon of S. enterica.jpg|center|frame| | + | [[Image:Eut Operon of S. enterica.jpg|center|frame|Figure 2. The 17 genes of the Eut operon. S, M, N, L, and K are believed to be components of the shell structure.]] |
| - | |[[Image:Eut BMC.jpg|frame| | + | |[[Image:Eut BMC.jpg|frame|Figure 3. Model of the Eut Shell structure.]] |
| - | |[[Image:Eut Metabolism.jpg|frame| | + | |[[Image:Eut Metabolism.jpg|frame|Figure 4. Model for the metabolism of ethanolamine in the Eut BMC. Ethanolamine enters the compartment which is believed to be composed of EutS, M, N, L, and K. It is then converted into acetaldehyde by EutBC. The compartment prevents acetaldehyde from diffusing away. EutG converts acetaldehyde to ethanol. EutE also converts trapped acetaldehyde into acetyl-CoA. This is then phosphorylated by EutD. Acetyl-phosphate and ethanol can then freely diffuse out of the compartment (adapted from Brinsmade et al. 2005)]] |
|} | |} | ||
| Line 32: | Line 33: | ||
| - | The five Eut proteins possessing BMC domains are the EutS, M, N, L, and K gene products. The first step in this project was to isolate these genes from genomic S. enterica DNA, specifically strain LT-2. Gene-specific primers containing 5' BglII and 3' NotI sites were designed for the five genes. EutL and EutK were cloned as a single PCR fragment, as were EutM and EutN. EutS was cloned separately. Each PCR product was ligated into the biobrick-compatible pUCBB plasmid (Johnson et al., manuscript in preparation). The EutMN construct was digested with EcoRI and SpeI and gel purified to prepare an entry fragment. The EutLK construct was linearized with EcoRI and XbaI. A EutMNLK construct was obtained by ligating the EutMN fragment into the linearized pUCBB-EutLK plasmid. This EutMNLK plasmid was then linearized with EcoRI/ XbaI and a final ligation was performed with EcoRI/SpeI digested EutS as the entry fragment. For submission to the iGEM competition the full EutSMNLK fragment was ligated into the psb1C3 vector. All completed constructs were confirmed by DNA sequencing. | + | The five Eut proteins possessing BMC domains are the EutS, M, N, L, and K gene products. The first step in this project was to isolate these genes from genomic S. enterica DNA, specifically strain LT-2. Gene-specific primers containing 5' BglII and 3' NotI sites were designed for the five genes. EutL and EutK were cloned as a single PCR fragment, as were EutM and EutN (Figure 5). EutS was cloned separately. Each PCR product was ligated into the biobrick-compatible pUCBB plasmid (Johnson et al., manuscript in preparation). The EutMN construct was digested with EcoRI and SpeI and gel purified to prepare an entry fragment. The EutLK construct was linearized with EcoRI and XbaI. A EutMNLK construct was obtained by ligating the EutMN fragment into the linearized pUCBB-EutLK plasmid. This EutMNLK plasmid was then linearized with EcoRI/ XbaI and a final ligation was performed with EcoRI/SpeI digested EutS as the entry fragment. For submission to the iGEM competition the full EutSMNLK fragment was ligated into the psb1C3 vector. All completed constructs were confirmed by DNA sequencing. |
| - | [[Image:Vectors.png|100px|frame| | + | [[Image:Vectors.png|100px|center|frame|Figure 5. Cloning and stacking of Eut BMC biobricks.]] |
| Line 39: | Line 40: | ||
To demonstrate that these genes can be expressed in a new bacterial host, these plasmids were transformed into E. coli and analysis of the protein produced was performed by SDS-PAGE. As shown in figures 6 and 7, our results indicate that E. coli are able to express the Eut BMC Genes. The observed overexpressed-protein bands match the predicted sizes of the recombinant proteins. pUCBB-GFP total cell lysate was used as a negative control in this experiment. | To demonstrate that these genes can be expressed in a new bacterial host, these plasmids were transformed into E. coli and analysis of the protein produced was performed by SDS-PAGE. As shown in figures 6 and 7, our results indicate that E. coli are able to express the Eut BMC Genes. The observed overexpressed-protein bands match the predicted sizes of the recombinant proteins. pUCBB-GFP total cell lysate was used as a negative control in this experiment. | ||
{| | {| | ||
| - | [[Image:Minnesota gel 4.png|center|frame| | + | [[Image:Minnesota gel 4.png|center|frame|Figure 6. SDS-PAGE gel showing recombinant Eut protein expression in E.coli]] |
| - | [[Image:Minnesota gel 5.png|center|frame| | + | [[Image:Minnesota gel 5.png|center|frame|Figure 7. SDS-PAGE gel showing expression of Eut SMNLK proteins from stacked biobricks. #5 and #6 are protein extracts from two independent E. coli colonies ]] |
|} | |} | ||
| Line 51: | Line 52: | ||
| - | Next, we wanted to determine which of the Eut BMC genes is required for localization of the tagged GFP reporter. As shown in Figure 8, co-transformations of EutMN, EutLK, and EutMNLK with the tagged reporter do not demonstrate the localizing phenomena. To our surprise, co-transformation with EutS alone was sufficient for localization! This observation indicates that EutS is the specific protein that recognizes the EutC signal sequence and causes localization of the tagged reporter. This discovery has caused us to become interested in investigating if EutS is able to form a proper BMC shell on its own. Our future experiments will probe further into this possibility — expect to hear more about this in next year’s jamboree! | + | Next, we wanted to determine which of the Eut BMC genes is required for localization of the tagged GFP reporter. As shown in Figure 8, co-transformations of EutMN, EutLK, and EutMNLK with the tagged reporter do not demonstrate the localizing phenomena. To our surprise, co-transformation of tagged GFP with EutS alone was sufficient for localization! This observation indicates that EutS is the specific protein that recognizes the EutC signal sequence and causes localization of the tagged reporter. This discovery has caused us to become interested in investigating if EutS is able to form a proper BMC shell on its own. Our future experiments will probe further into this possibility — expect to hear more about this in next year’s jamboree! |
== Discussion == | == Discussion == | ||
| - | [[Image:Eutc-ss_tag.gif|frame| | + | [[Image:Eutc-ss_tag.gif|frame|Figure 9. Model for compartmentalization of EutC tagged protein inside a properly formed BMC shell]] |
| - | We are pleased to show that our fluorescent images yielded such definite results. For many of us this was our first exposure to cloning experiments. The work performed at the outset of this project went smoothly, but by the end of the summer we were facing | + | We are pleased to show that our fluorescent images yielded such definite results. For many of us this was our first exposure to cloning experiments. The work performed at the outset of this project (cloning and expression of Eut genes) went smoothly, but by the end of the summer we were facing setbacks in our attempts to clone enzymes to introduce into our BMCs. However, we are satisfied that our results have potentially valuable applications. An example application would be to tag enzymes with the EutC signal sequence and co-transform these with EutS. The enzymes would associate to a definite location in the cytoplasm and biocatalysis would be improved by bringing multiple steps of a pathway into proximity. We hope to see future iGEM teams develop imaginative new uses for our Biobrick submission. |
| - | + | ||
| - | + | ||
| - | + | ||
| + | To move forward with our project, our goal will be to demonstrate that these proteins are forming a structured microcompartment and not a cytoplasmic agglutination (Figure 9). Further imaging experiments will be needed. The next step that our group will take in exploring BMCs and their properties will make use of scanning and transmission electron microscopes to visualize whether they form the hypothesized icosahedral structure. Other future experiments will attempt to show that this system can localize enzymes to engineer pathways with improved efficiency as well as prevent the accumulation of toxic intermediate in the cytoplasm, allowing cells to perform reactions that would otherwise result in their death. Nanobioreactors! | ||
== References == | == References == | ||
Latest revision as of 15:33, 27 October 2010
| Home | Team | Project | Protocols | Notebook | Judging Criteria | Attributions | Safety |
|---|
Contents |
Introduction
A bacterial microcompartment (BMC) is a polyhedral protein complex that encapsulates enzymes involved in specific metabolic pathways. BMCs were first observed by microscope in cyanobacteria the 1950’s. By the late 1990’s, it had become clear that these proteinaceous structures are produced by many types of bacteria, and serve various functions (Yeates et al. 2008). There are multiple examples of BMC’s occurring in nature: the carboxysome found in cyanobacteria and chemoautotrophic bacteria, the propanediol utilization compartment found in enterobacteria, as well as the ethanolamine utilization (Eut) compartment used in this project, also found in enterobacteria. By sequestering enzymes and substrates together in an enclosed space within the cell, the overall reaction efficiency is greatly improved. In some species, BMCs localize toxic compounds with enzymes that convert them into benign compounds. In this way a bacterial cell can ensure that the problematic intermediate is not released into the cell cytoplasm.
BMCs are ideal for iGEM because they could be harnessed as nanobioreactors for metabolic engineering. Interest in BMCs and their natural functions has grown in recent years. With increasing availability of information of their properties, BMCs are becoming more practical for applications in synthetic biology. Engineering a BMC to house an enzymatic pathway would increase the efficiency of the process, and would also quarantine any toxic intermediates (Figure 1).
Our goals were to develop E. coli that express a recombinant BMC, and develop a system for targeting proteins to be encapsulated within that compartment. We chose to use the Ethanolamine utilization (Eut) shell from Salmonella enterica because this species has been shown to produce these structures and they have been well characterized (Penrod & Roth, 2006). The ethanolamine utilization operon from Salmonella enterica strain LT2 is composed of 17 genes (Figure 2). Of these, only five possess BMC associated domains and are believed to be components of the Eut microcompartment (Figure 3). In Salmonella enterica this organelle-like structure localizes the enzymes required for ethanolamine utilization and prevents acetaldehyde, a volatile intermediate, from diffusing into the cytoplasm (Figure 4).
Results
The five Eut proteins possessing BMC domains are the EutS, M, N, L, and K gene products. The first step in this project was to isolate these genes from genomic S. enterica DNA, specifically strain LT-2. Gene-specific primers containing 5' BglII and 3' NotI sites were designed for the five genes. EutL and EutK were cloned as a single PCR fragment, as were EutM and EutN (Figure 5). EutS was cloned separately. Each PCR product was ligated into the biobrick-compatible pUCBB plasmid (Johnson et al., manuscript in preparation). The EutMN construct was digested with EcoRI and SpeI and gel purified to prepare an entry fragment. The EutLK construct was linearized with EcoRI and XbaI. A EutMNLK construct was obtained by ligating the EutMN fragment into the linearized pUCBB-EutLK plasmid. This EutMNLK plasmid was then linearized with EcoRI/ XbaI and a final ligation was performed with EcoRI/SpeI digested EutS as the entry fragment. For submission to the iGEM competition the full EutSMNLK fragment was ligated into the psb1C3 vector. All completed constructs were confirmed by DNA sequencing.
To demonstrate that these genes can be expressed in a new bacterial host, these plasmids were transformed into E. coli and analysis of the protein produced was performed by SDS-PAGE. As shown in figures 6 and 7, our results indicate that E. coli are able to express the Eut BMC Genes. The observed overexpressed-protein bands match the predicted sizes of the recombinant proteins. pUCBB-GFP total cell lysate was used as a negative control in this experiment.
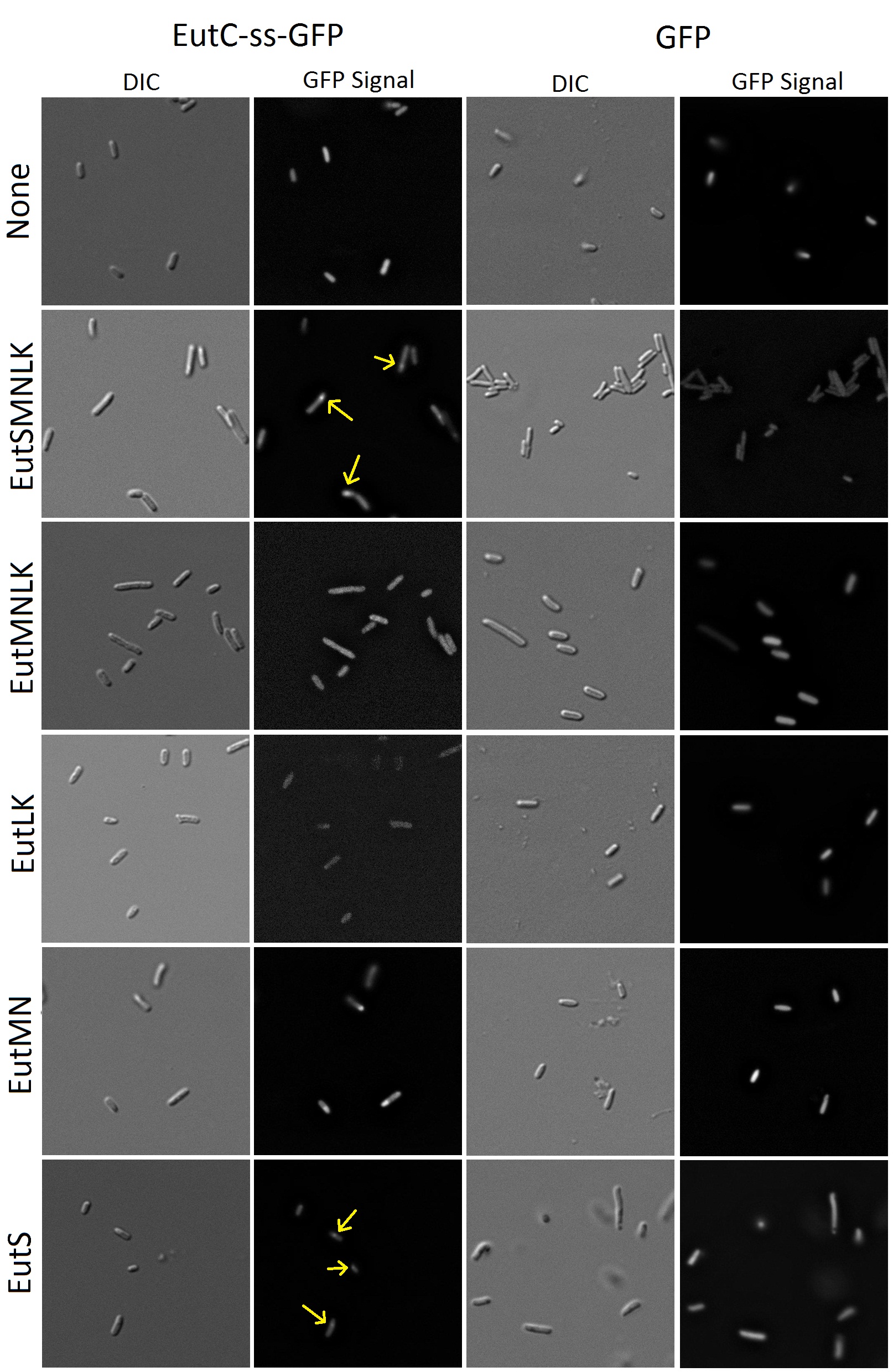
To a synthetic biologist, an empty proteinacious compartment by itself is not useful without a way of targeting favorite proteins to the inside. It had been demonstrated that signal sequences at the N-terminus of BMC associated enzymes are capable of causing their compartmentalization inside shells (Cheng et al.). To be able to target a reporter to the inside of our recombinant Eut BMCs, our group used a predicted signal sequence from the enzyme EutC, which is known to be sequestered inside the Eut BMC. EutC encodes the small subunit of the ethanolamine ammonia lyase enzyme that is known to be targeted to the Eut microcompartment. By comparison to homologous enzymes, Cheng et al. suggested that a sequence of 19 amino acids at the N-terminus of EutC may be a likely candidate for serving this function. To verify this idea, a construct containing this signal sequence fused to GFP was developed and co-transformed in E. coli cells alongside the Eut SMNLK vector. These dual-transformed cells were characterized by fluorescence microscopy and the results are summarized below in figure 8.
Figure 8. Fluorescence and DIC images of E.coli cells transformed with GFP (with or without EutC signal sequence)and Eut BMC genes.
Flourescence imaging shows that cells transformed solely with GFP or GFP with the fused signal sequence cause the entirety of the cell cytoplasm to fluoresce. The cells transformed with untagged-GFP and the microcompartment proteins also have distributed fluorescence. We were very happy to find that E. coli transformed with tagged GFP and all of the microcompartment genes (EutSMNLK) display localization of the fluorescence to a discrete point within the cell. This suggests that recombinant EutSMNLK are indeed able to form a BMC within E. coli, and the tagged GFP reporter localizes to these BMCs. These experiments have shown that the S. enterica microcompartment proteins can be expressed in E. coli and that expression of these proteins can cause GFP with an attached signal sequence to be localized to a distinct point inside the cell.
Next, we wanted to determine which of the Eut BMC genes is required for localization of the tagged GFP reporter. As shown in Figure 8, co-transformations of EutMN, EutLK, and EutMNLK with the tagged reporter do not demonstrate the localizing phenomena. To our surprise, co-transformation of tagged GFP with EutS alone was sufficient for localization! This observation indicates that EutS is the specific protein that recognizes the EutC signal sequence and causes localization of the tagged reporter. This discovery has caused us to become interested in investigating if EutS is able to form a proper BMC shell on its own. Our future experiments will probe further into this possibility — expect to hear more about this in next year’s jamboree!
Discussion
We are pleased to show that our fluorescent images yielded such definite results. For many of us this was our first exposure to cloning experiments. The work performed at the outset of this project (cloning and expression of Eut genes) went smoothly, but by the end of the summer we were facing setbacks in our attempts to clone enzymes to introduce into our BMCs. However, we are satisfied that our results have potentially valuable applications. An example application would be to tag enzymes with the EutC signal sequence and co-transform these with EutS. The enzymes would associate to a definite location in the cytoplasm and biocatalysis would be improved by bringing multiple steps of a pathway into proximity. We hope to see future iGEM teams develop imaginative new uses for our Biobrick submission.
To move forward with our project, our goal will be to demonstrate that these proteins are forming a structured microcompartment and not a cytoplasmic agglutination (Figure 9). Further imaging experiments will be needed. The next step that our group will take in exploring BMCs and their properties will make use of scanning and transmission electron microscopes to visualize whether they form the hypothesized icosahedral structure. Other future experiments will attempt to show that this system can localize enzymes to engineer pathways with improved efficiency as well as prevent the accumulation of toxic intermediate in the cytoplasm, allowing cells to perform reactions that would otherwise result in their death. Nanobioreactors!
References
Fan, C. G., S. Q. Cheng, et al. (2010). "Short N-terminal sequences package proteins into bacterial microcompartments." Proceedings of the National Academy of Sciences of the United States of America 107(16): 7509-7514.
Penrod, J. T. and J. R. Roth (2006). "Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica." Journal of Bacteriology 188(8): 2865-2874.
Yeates, T. O., C. A. Kerfeld, et al. (2008). "Protein-based organelles in bacteria: carboxysomes and related microcompartments." Nature Reviews Microbiology 6(9): 681-691.
 "
"