Team:Heidelberg/Notebook/miRNA Kit/September
From 2010.igem.org
(→29/09/2010) |
|||
| (92 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Pagetop|note_mirna_kit}} |
| + | {{:Team:Heidelberg/Side_Top}} | ||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#009be1; border: 1.5px solid #000000;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#f09600;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August<font color="white">August</font>] | ||
| + | |- style="background:#f09600; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |colspan="6"| ||'''1''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''2'''||'''3'''||'''4'''||'''5'''||'''6'''||'''7'''||'''8''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''9'''||'''10'''||'''11'''||'''12'''||'''13'''||'''14'''||'''15''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''16'''||'''17'''||'''18'''||'''19'''||'''20'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#21/08/2010 21]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#22.2F08.2F2010 22]''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#23.2F08.2F2010 23]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#24.2F08.2F2010 24]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#25.2F08.2F2010 25]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#26.2F08.2F2010 26]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#27.2F08.2F2010 27]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#28.2F08.2F2010 28]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/August#29.2F08.2F2010 29]''' | ||
| + | |- style="background:#f2f2f2; color:#009be1" | ||
| + | |'''[[Igem2010/Main/synthetic_miR_Kit/August#30/08/2010|30]]'''||'''[[Igem2010/Main/synthetic_miR_Kit/August#31/08/2010|31]]'''||colspan="5"| | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#78b41e; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#009be1;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September<font color="#ffecba">September</font>] | ||
| + | |- style="background:#009be1; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="2"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#01.2F09.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#02.2F09.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#03.2F09.2F2010 3]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#04.2F09.2F2010 4]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#05.2F09.2F2010 5]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#06.2F09.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#07.2F09.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#08.2F09.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#09.2F09.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#10.2F09.2F2010 10]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#11.2F09.2F2010 11]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#12.2F09.2F2010 12]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#13.2F09.2F2010 13]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#14.2F09.2F2010 14]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#15.2F09.2F2010 15]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#16.2F09.2F2010 16]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#17.2F09.2F2010 17]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#18.2F09.2F2010 18]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#19.2F09.2F2010 19]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#20.2F09.2F2010 20]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#21.2F09.2F2010 21]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#22.2F09.2F2010 22]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#23.2F09.2F2010 23]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#24.2F09.2F2010 24]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#25.2F09.2F2010 25]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#26.2F09.2F2010 26]''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#27.2F09.2F2010 27]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#28.2F09.2F2010 28]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#29.2F09.2F2010 29]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#22.2F30.2F2010 30]'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| cellpadding="5" cellspacing="0" align="center" style="text-align: center; color:#f09600; border: 1.5px solid #333333;" | ||
| + | |- border="0" | ||
| + | ! colspan="7" style="background:#78b41e;" | [https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October<font color="white">October</font>] | ||
| + | |- style="background:#78b41e; color:white" | ||
| + | |width="20pt"|'''M'''||width="20pt"|'''T'''||width="20pt"|'''W'''||width="20pt"|'''T'''||width="20pt"|'''F'''||width="20pt"|'''S'''||width="20pt"|'''S''' | ||
| + | |- style="background:#f2f2f2; color:#78b41e" | ||
| + | |colspan="4"| ||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#01.2F10.2F2010 1]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#02.2F10.2F2010 2]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#03.2F10.2F2010 3]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#04.2F10.2F2010 4]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#05.2F10.2F2010 5]''' | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#06.2F10.2F2010 6]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#07.2F10.2F2010 7]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#08.2F10.2F2010 8]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#09.2F10.2F2010 9]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#10.2F10.2F2010 10]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#11.2F10.2F2010 11]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#12.2F10.2F2010 12]'''|'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#13.2F10.2F2010 13]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#14.2F10.2F2010 14]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#15.2F09.2F2010 15]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#16.2F10.2F2010 16]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#17.2F10.2F2010 17]'''||'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#18.2F10.2F2010 18]''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/October#19/10/2010 19]'''||'''20'''||'''21'''||'''22'''||'''23'''||'''24'''||'''25''' | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | |'''26'''||'''27'''||'''28'''||'''29'''||'''30'''||colspan="3"| | ||
| + | |- style="background:#f2f2f2; color:#f09600" | ||
| + | | colspan="7"| <span style="color:#ffffff">-</span> | ||
| + | |} | ||
| + | |||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
| + | |||
| + | __NOTOC__ | ||
===01/09/2010=== | ===01/09/2010=== | ||
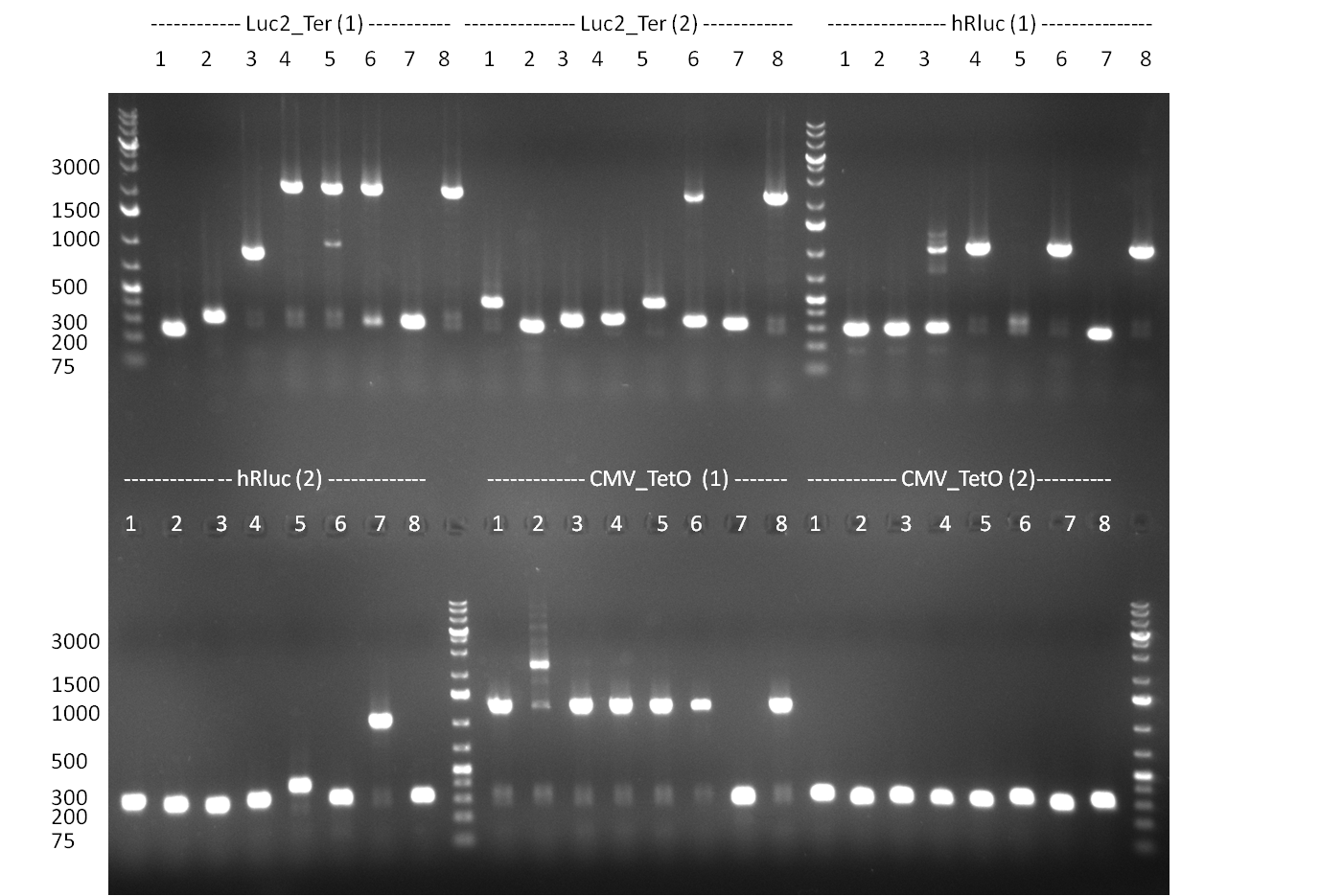
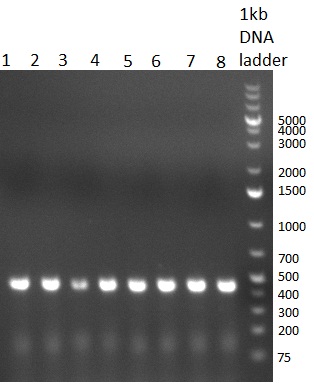
| - | [[Image:20100901_gel1bb.png|thumb|350 px| | + | [[Image:20100901_gel1bb.png|thumb|350 px|left|Gel 100901-1]] |
| - | [[Image:20100901_gel2bb.png|thumb|350 px| | + | [[Image:20100901_gel2bb.png|thumb|350 px|left|Gel 100901-2]] |
<br /> | <br /> | ||
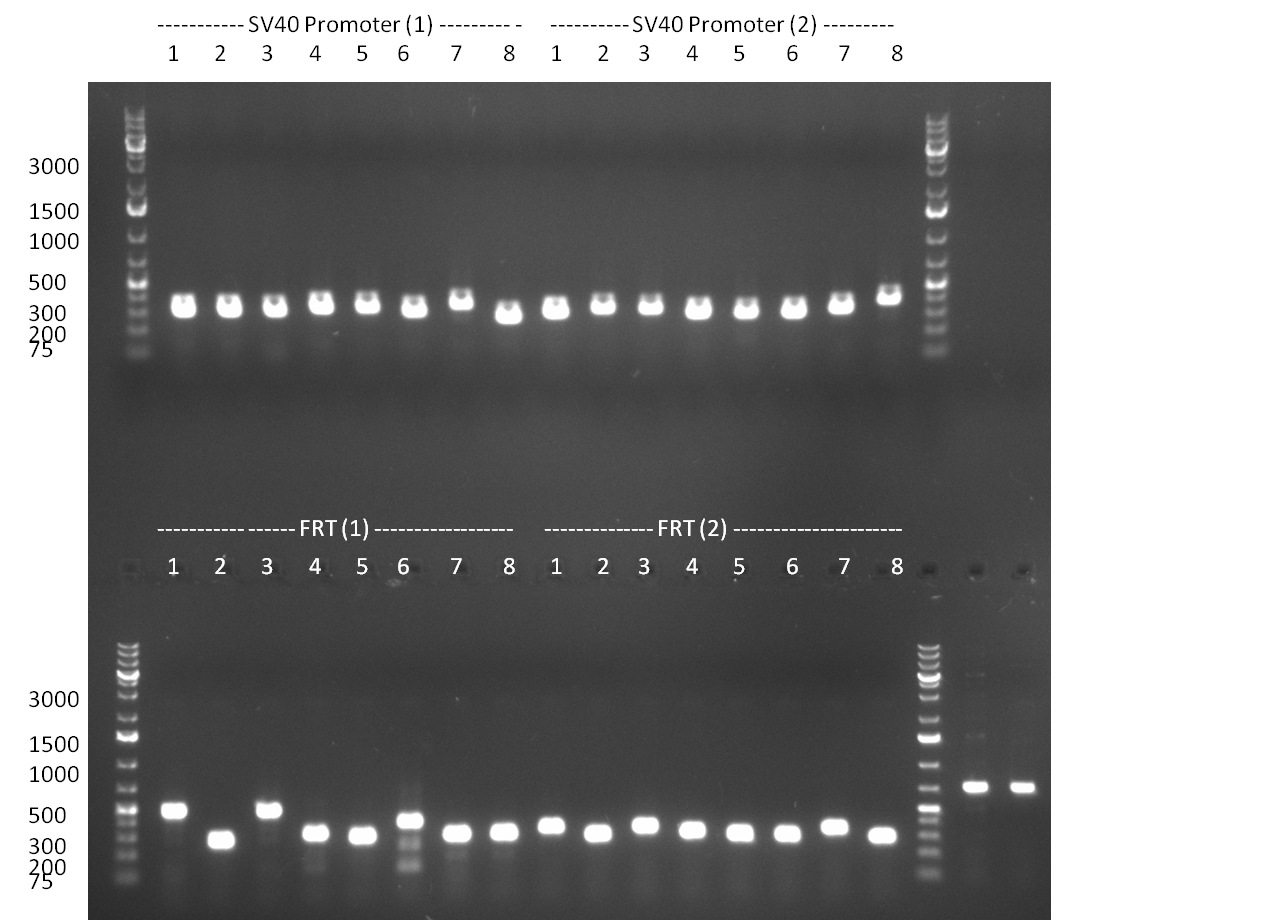
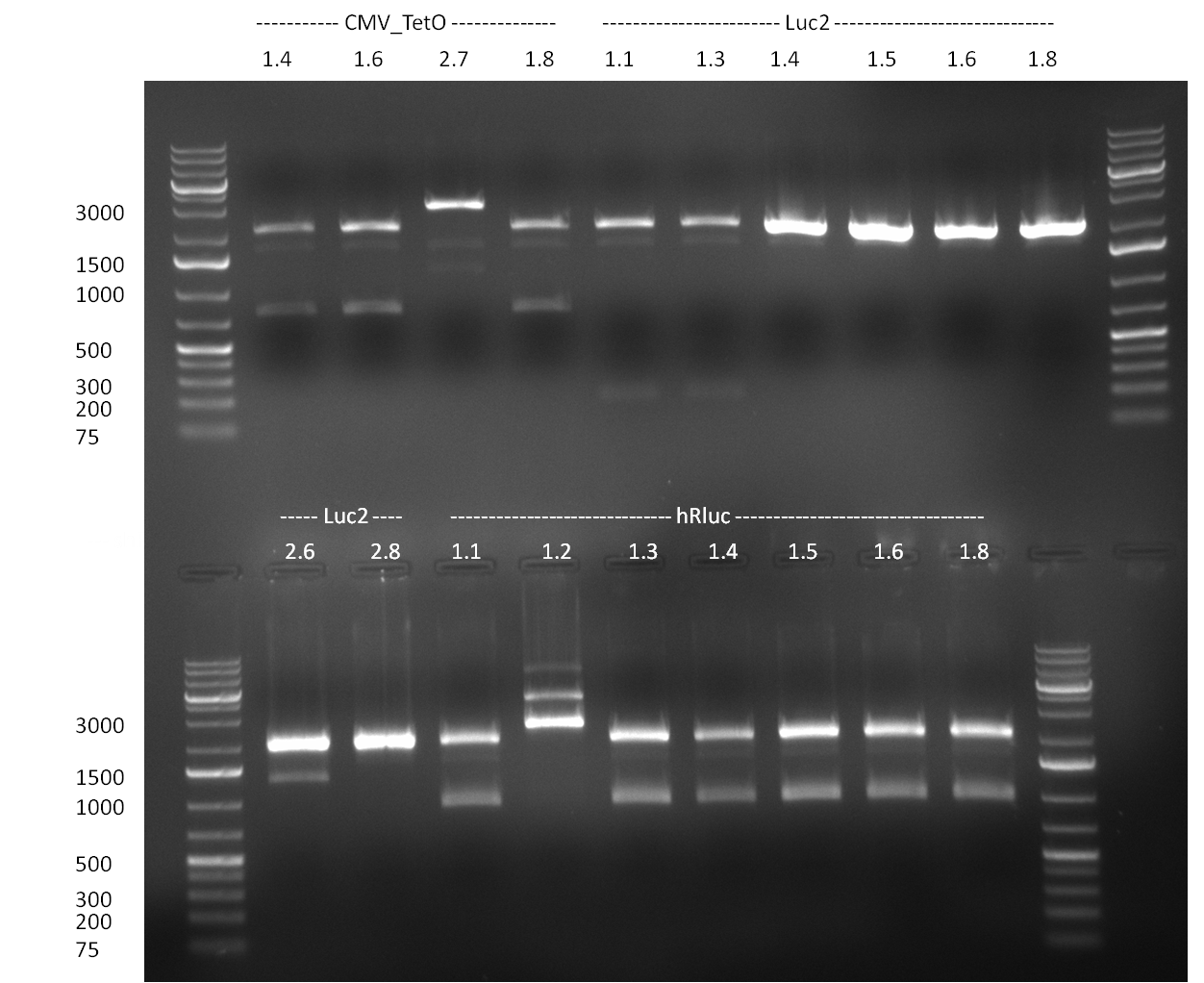
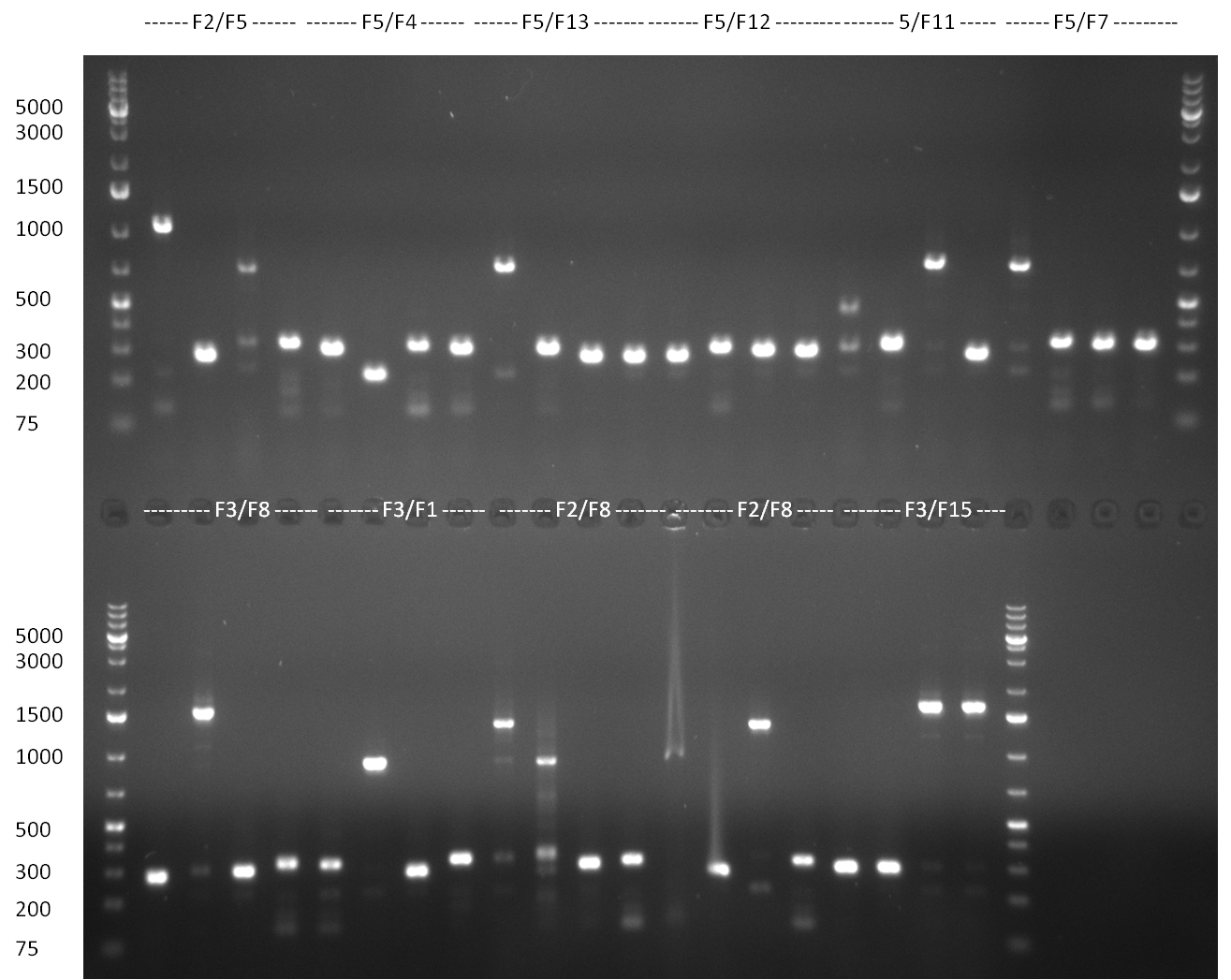
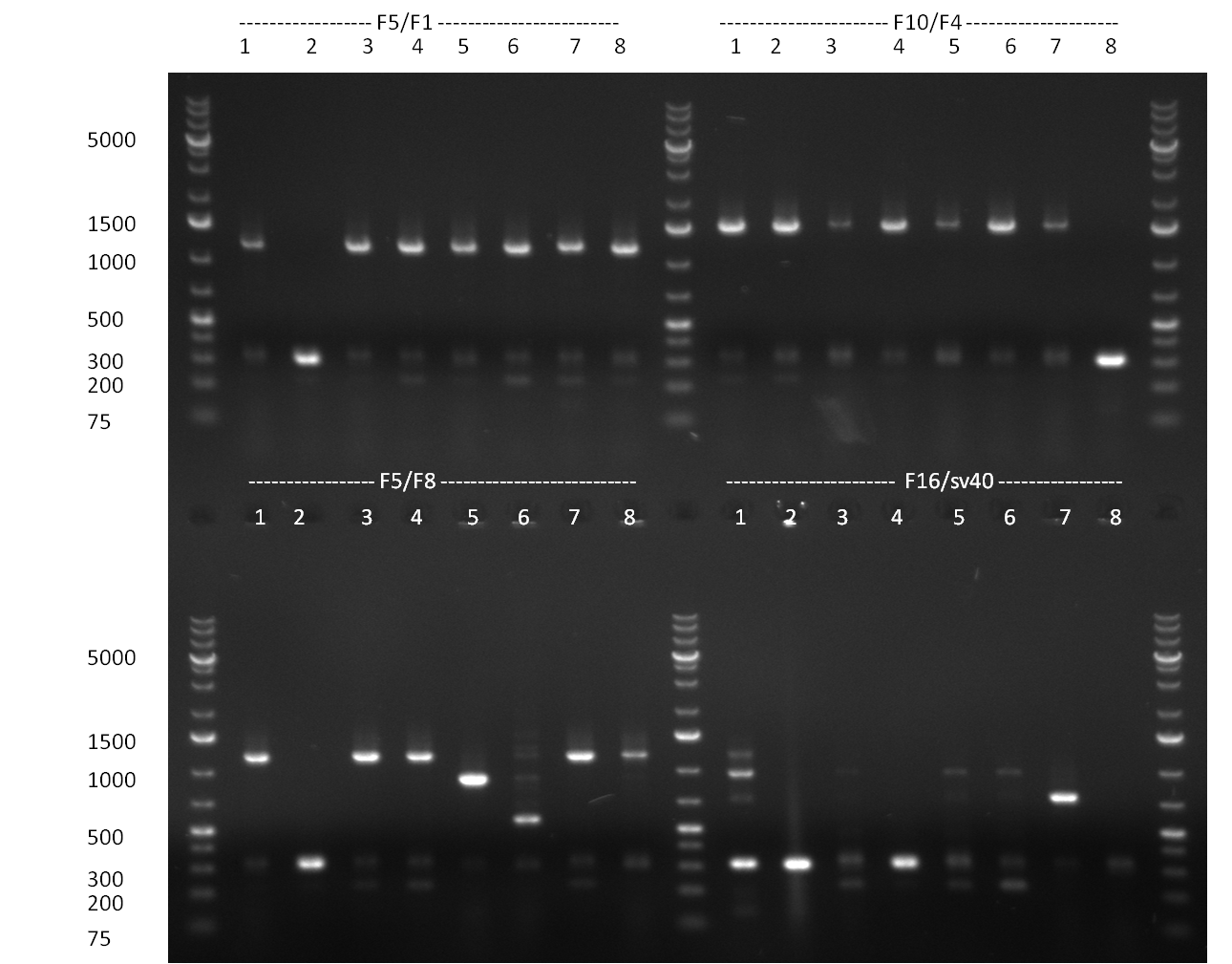
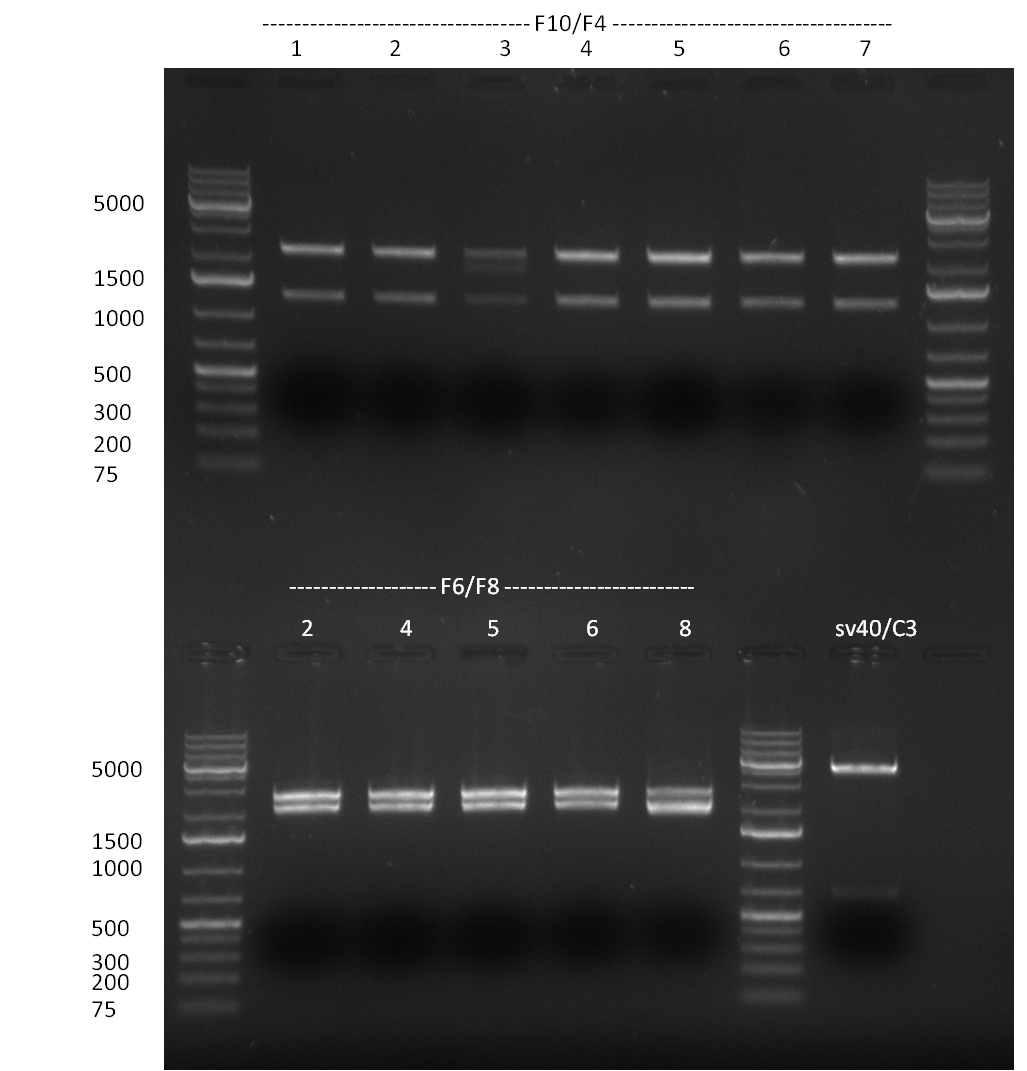
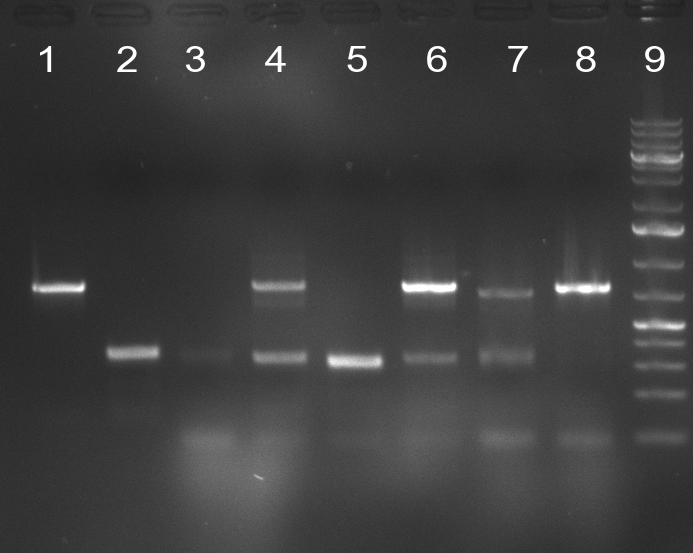
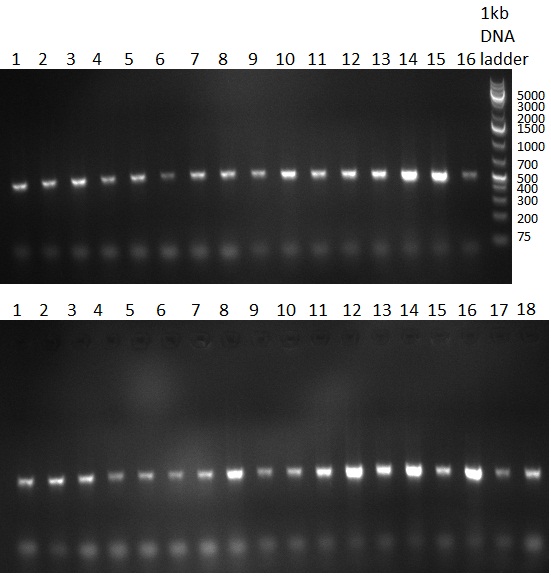
* 8 colonies of each plate (see [[Igem2010/Main/synthetic miR Kit/August#31/08/2010 | previous day]]) were picked and colony-PCR was performed. Furthermore, the colonies were used for inocculation of LB miniprep cultures. The numbers behind the construct name refer to vector type (1 = not SAP treated, 2 = dephosphorylated using SAP; see previous day). For hRluc (1300 bp band)), Luc2 (2300 bp band), CMV_TetO (1000 bp band) and the FRT (500 bp band) site, positive colonies could be observed (gel 100901-1 and 100901-2). | * 8 colonies of each plate (see [[Igem2010/Main/synthetic miR Kit/August#31/08/2010 | previous day]]) were picked and colony-PCR was performed. Furthermore, the colonies were used for inocculation of LB miniprep cultures. The numbers behind the construct name refer to vector type (1 = not SAP treated, 2 = dephosphorylated using SAP; see previous day). For hRluc (1300 bp band)), Luc2 (2300 bp band), CMV_TetO (1000 bp band) and the FRT (500 bp band) site, positive colonies could be observed (gel 100901-1 and 100901-2). | ||
| Line 12: | Line 77: | ||
<br /> | <br /> | ||
===02/09/2010=== | ===02/09/2010=== | ||
| - | [[Image:20100802_gel1.png|thumb|350 px| | + | [[Image:20100802_gel1.png|thumb|350 px|left|Gel 100902-1]] |
| + | [[Image:20100902_gel2.png|thumb|350 px|right|Gel 100902-2]] | ||
| + | [[Image:20100902_gel3.png|thumb|350 px|right|Gel 100902-3]] | ||
<br /> | <br /> | ||
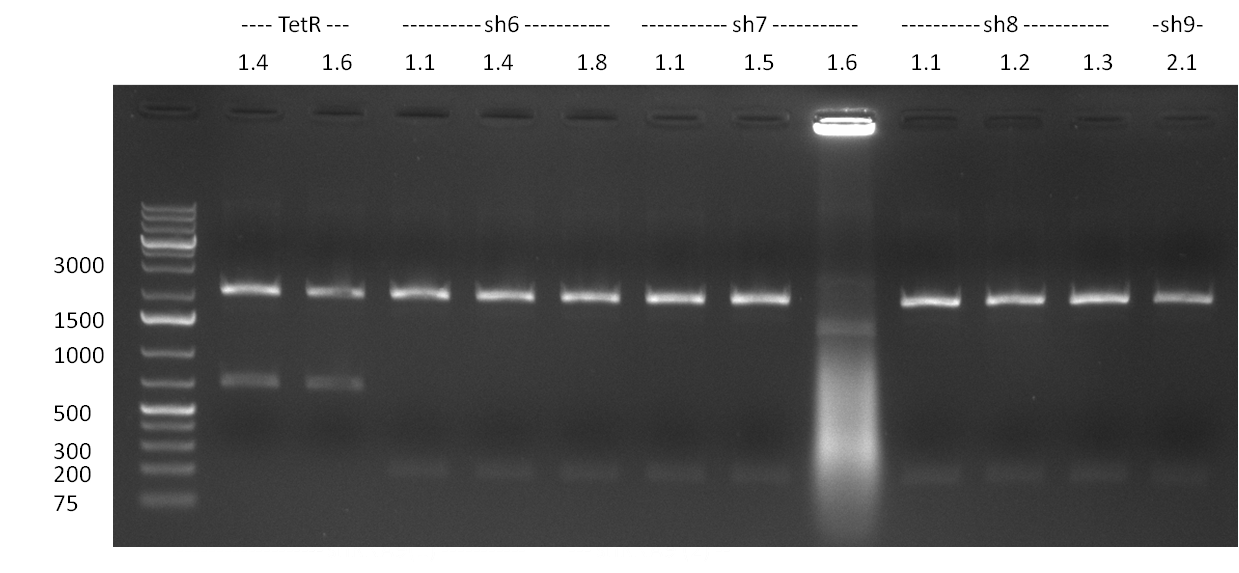
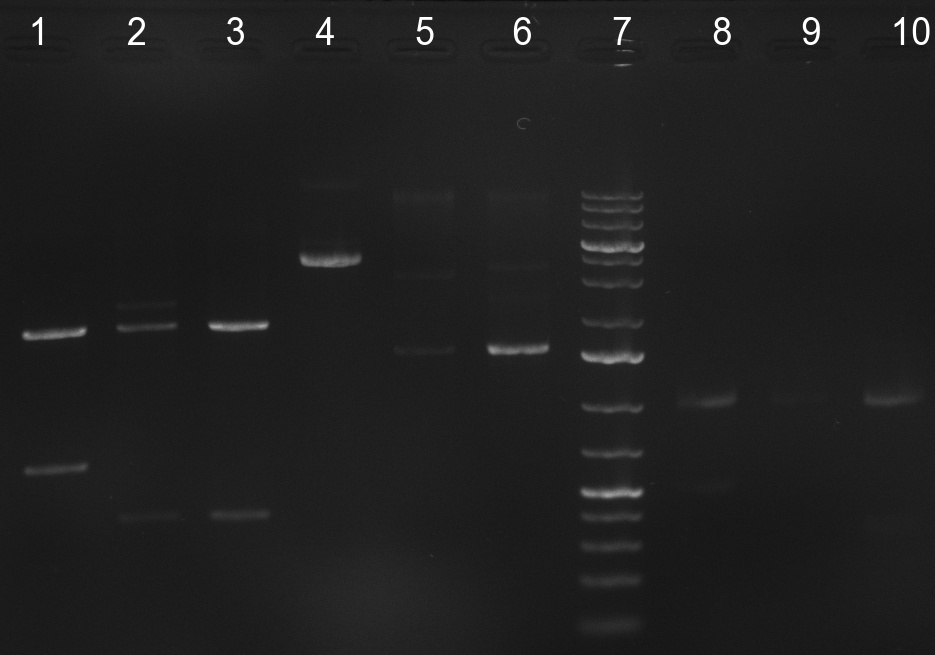
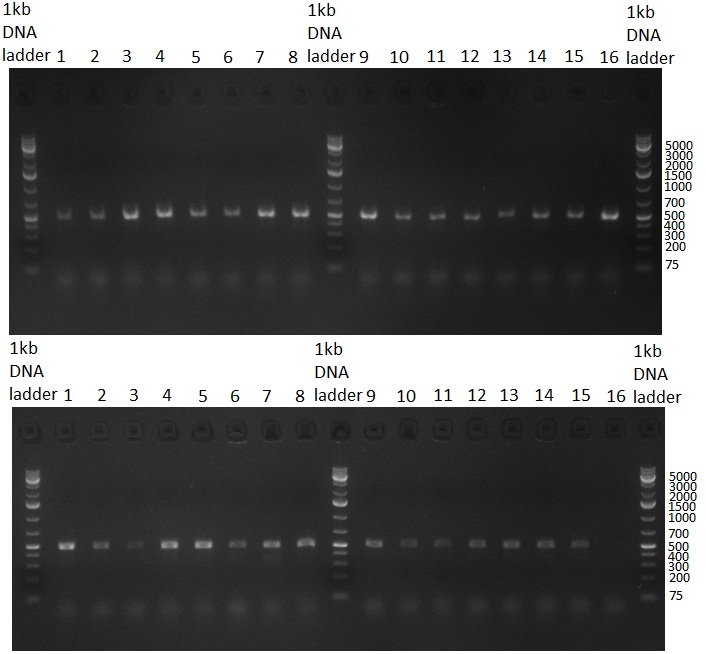
| - | * 8 colonies were picked from plates 1 and 4 colonies from plates 2 from the previous days cloning and analyzed via colony PCR performed according to the [ | + | * 8 colonies were picked from plates 1 and 4 colonies from plates 2 from the previous days cloning and analyzed via colony PCR performed according to the [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Colony_PCR PCR standard protocol]. The results of the colony PCR were analyzed on a 1 % agarose gel run for 1 h @ 100 V (gel 100902-1 and 100902-2). The following colonies were afterwards inocculated (LB miniprep cultures): |
:* TetR (1.4 and 1.6) | :* TetR (1.4 and 1.6) | ||
:* shRNA6 (1.1, 1.4 and 1.8) | :* shRNA6 (1.1, 1.4 and 1.8) | ||
| Line 21: | Line 88: | ||
:* shRNA9 (1.2) | :* shRNA9 (1.2) | ||
<br /> | <br /> | ||
| + | |||
* The cultures inocculated the day before were mini prepped by applying a Qiagen Miniprep Kit. Afterwards, a test digestion with NotI was performed and the result was analyzed on a 1 % agarose gel, run for 35 min @ 100 V (gel 100902-3). According to the test digestion result, the following samples were send for sequencing @ GATC: | * The cultures inocculated the day before were mini prepped by applying a Qiagen Miniprep Kit. Afterwards, a test digestion with NotI was performed and the result was analyzed on a 1 % agarose gel, run for 35 min @ 100 V (gel 100902-3). According to the test digestion result, the following samples were send for sequencing @ GATC: | ||
:* CMV_TetO2 (.4, 1.6) | :* CMV_TetO2 (.4, 1.6) | ||
| Line 27: | Line 95: | ||
:* hRluc (1.1, 1.3, 1.4) | :* hRluc (1.1, 1.3, 1.4) | ||
<br /> | <br /> | ||
| - | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
===03/09/2010=== | ===03/09/2010=== | ||
| Line 43: | Line 109: | ||
* cloning of shRNAs into pcDNA5/FRT/TO | * cloning of shRNAs into pcDNA5/FRT/TO | ||
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
| - | + | <br /><br /><br /> | |
===04/09/2010=== | ===04/09/2010=== | ||
| Line 109: | Line 175: | ||
* ligation of both inserts for the tuning construct, extraction of the right insert (3000bp) and ligation into the backbone | * ligation of both inserts for the tuning construct, extraction of the right insert (3000bp) and ligation into the backbone | ||
| - | <br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /> |
===09/09/2010=== | ===09/09/2010=== | ||
| Line 121: | Line 187: | ||
<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
<br /><br /> | <br /><br /> | ||
| + | <br /><br /><br /><br /><br /><br /><br /><br /> | ||
| + | <br /><br /><br /><br /><br /> | ||
<br /><br /> | <br /><br /> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===10/09/2010=== | ===10/09/2010=== | ||
<br /> | <br /> | ||
| + | [[Image:100910-rbtettestdigest.jpg|thumb|350 px|right|Tet test digest ]] | ||
| + | [[Image:Rb100910colonyshRNAs.jpg|thumb|350 px|right| colony PCR shRNAs]] | ||
* PCR of SV40 Terminator with BamHI site according to standard PCR protocol (annealing @ 58 °C) | * PCR of SV40 Terminator with BamHI site according to standard PCR protocol (annealing @ 58 °C) | ||
* Assembly of the following constructs, again via 3A standard assembly. | * Assembly of the following constructs, again via 3A standard assembly. | ||
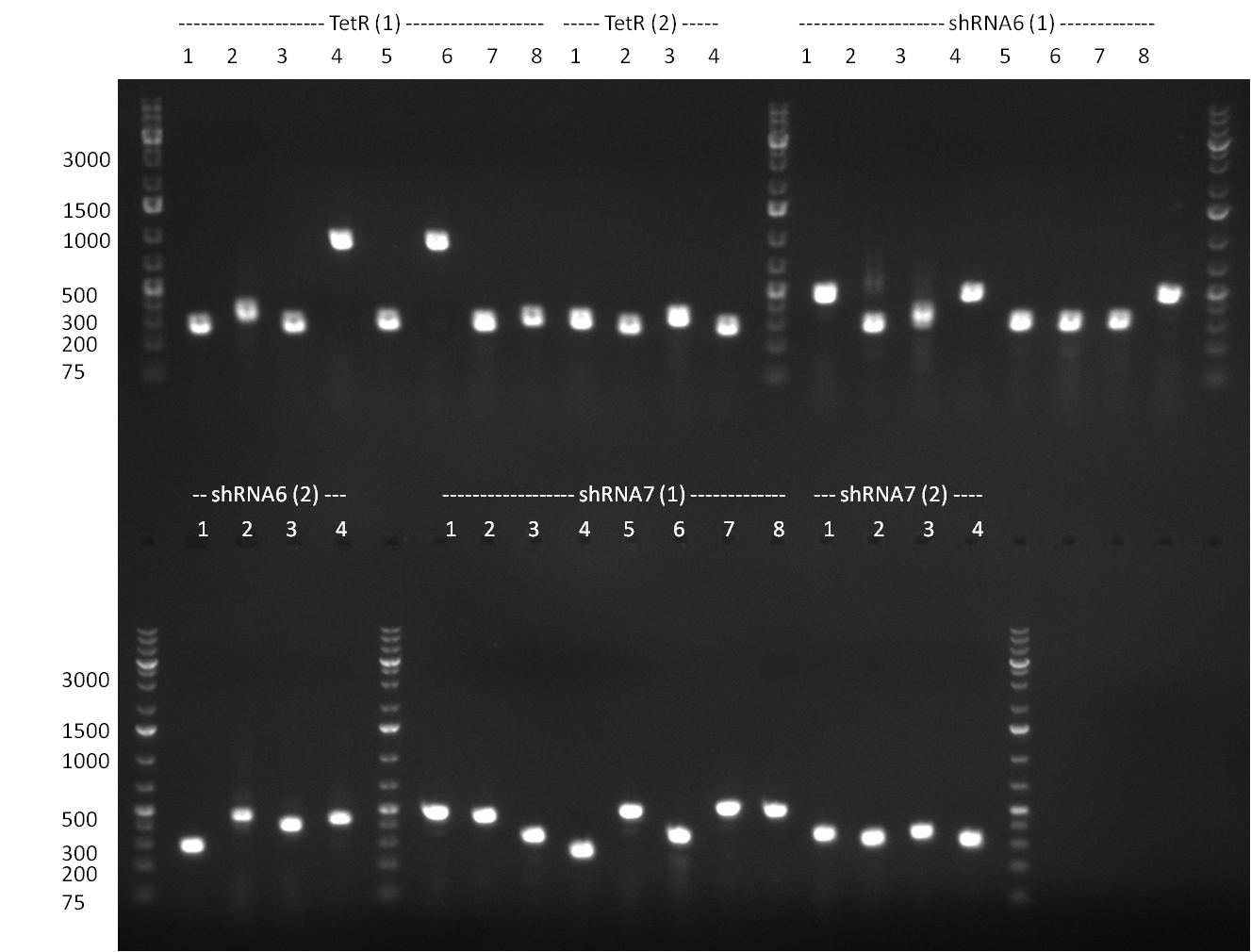
| Line 139: | Line 206: | ||
*** for lanes 2-4 you see a stronger band at 700 pb (size of TetR) | *** for lanes 2-4 you see a stronger band at 700 pb (size of TetR) | ||
*** for landes 5-7 you see a stronger band at 5000 bp as it was linearized with SgfI | *** for landes 5-7 you see a stronger band at 5000 bp as it was linearized with SgfI | ||
| - | + | ||
* colony PCR of binding site in PsiCheck and tuning construct.. tuning construct looks promising, BS nothing on the gel except for primers | * colony PCR of binding site in PsiCheck and tuning construct.. tuning construct looks promising, BS nothing on the gel except for primers | ||
* colony PCR of shRNAs look promising even though we cannot say whether still the old shRNA is inside | * colony PCR of shRNAs look promising even though we cannot say whether still the old shRNA is inside | ||
** PCR of CMV shRNA which leads to a size of 1200bp | ** PCR of CMV shRNA which leads to a size of 1200bp | ||
| - | |||
| + | <br /><br /><br /><br /><br /><br /><br /><br /><br /> | ||
===11/09/2010=== | ===11/09/2010=== | ||
| Line 155: | Line 222: | ||
* Tet Repressor cloned!!! | * Tet Repressor cloned!!! | ||
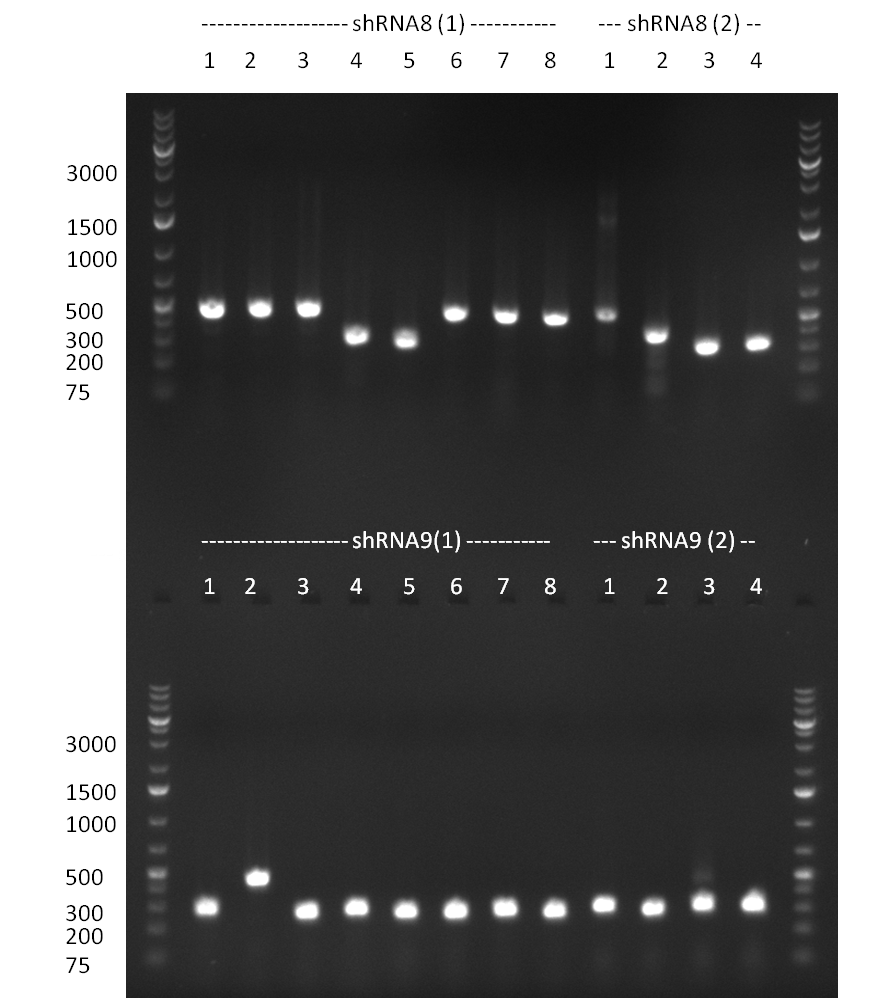
* shRNA 2,3,5,7,8 and 9 seems to be cloned according to test digest | * shRNA 2,3,5,7,8 and 9 seems to be cloned according to test digest | ||
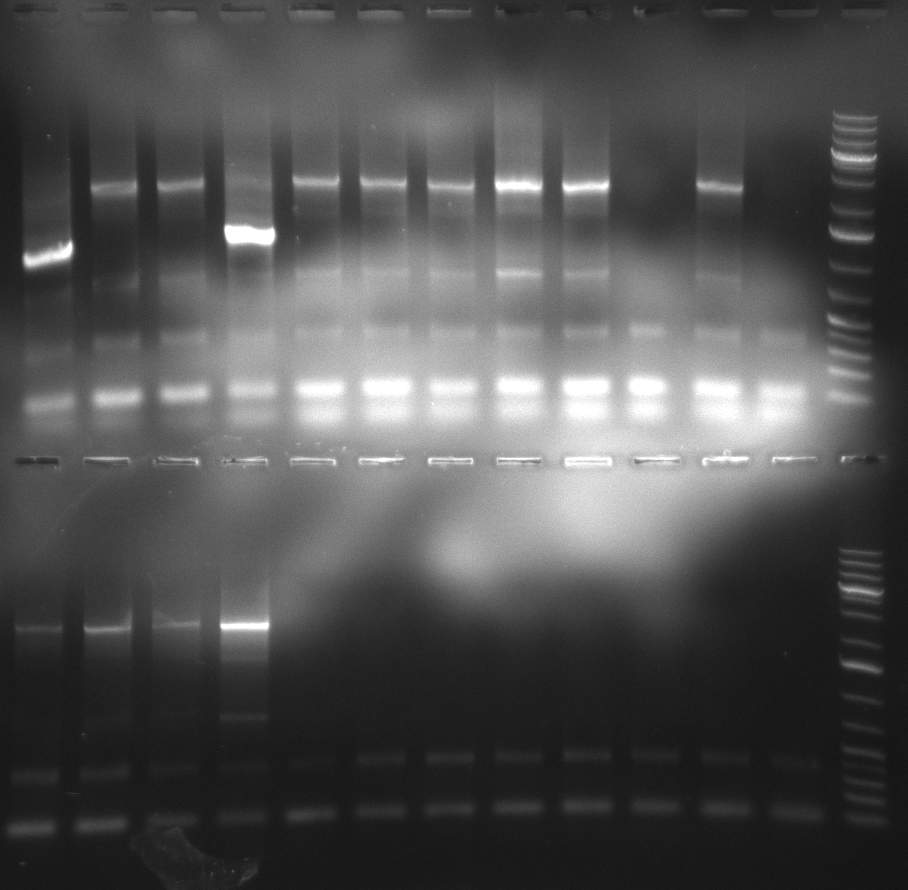
| - | [[Image:Rb100911_testdigestshRNA.jpg|350px]] | + | [[Image:Rb100911_testdigestshRNA.jpg|thumb|350px|right|test digest]] |
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
===12/09/2010=== | ===12/09/2010=== | ||
| Line 167: | Line 234: | ||
* miniprep of shRNA constructs and Tuning constructs | * miniprep of shRNA constructs and Tuning constructs | ||
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
===13/09/2010=== | ===13/09/2010=== | ||
| Line 194: | Line 261: | ||
* Colony- PCR for the cloning products done on the previous day (gels 100914-1 and 100914-2 on the right) | * Colony- PCR for the cloning products done on the previous day (gels 100914-1 and 100914-2 on the right) | ||
* positive clones were Mini-Prepped and test digested | * positive clones were Mini-Prepped and test digested | ||
| - | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | |
===15/09/2010=== | ===15/09/2010=== | ||
| Line 248: | Line 315: | ||
4 °C/ forever <br /> | 4 °C/ forever <br /> | ||
................................................ <br /> | ................................................ <br /> | ||
| - | + | ||
Each PCR was performed in two replicates. The result of the PCRs was analyzed on a 1 % agarose gel (100915-1), run for 1 h @ 100 V. The PCR nr. 2 was subsequently PCR purified by applying a Qiagen PCR purification KIT and used for the cloning | Each PCR was performed in two replicates. The result of the PCRs was analyzed on a 1 % agarose gel (100915-1), run for 1 h @ 100 V. The PCR nr. 2 was subsequently PCR purified by applying a Qiagen PCR purification KIT and used for the cloning | ||
| Line 278: | Line 345: | ||
<br /> | <br /> | ||
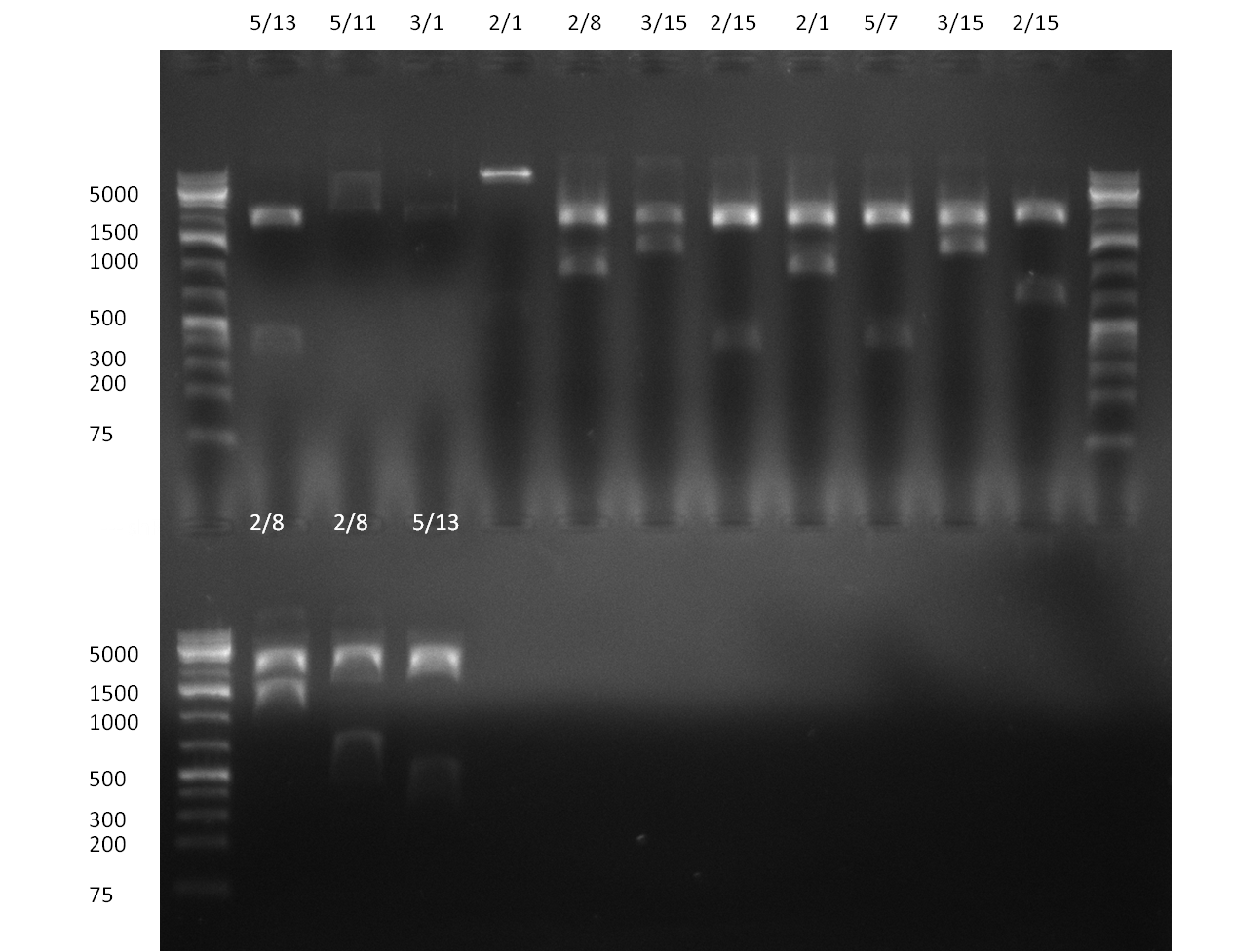
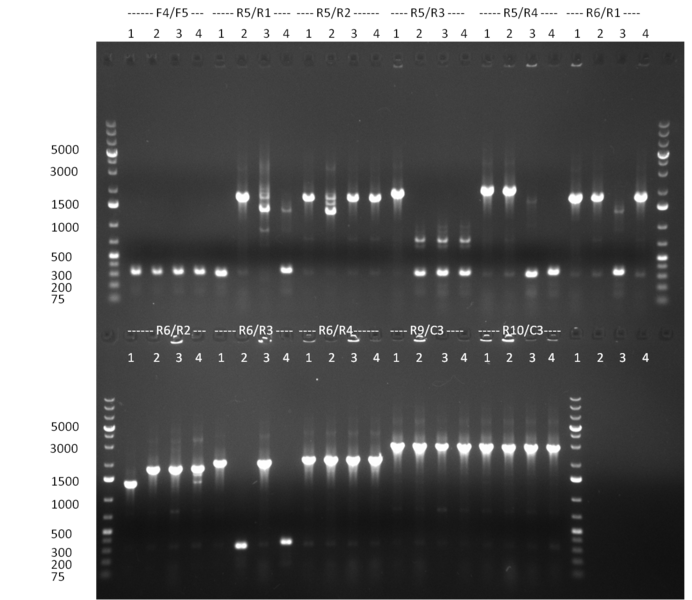
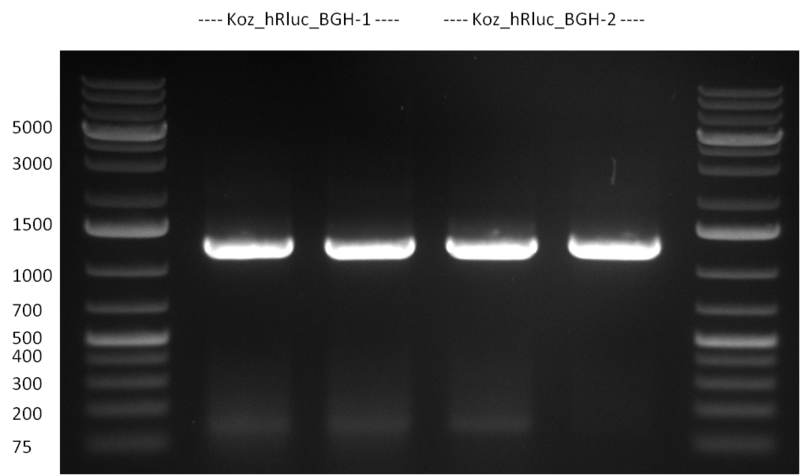
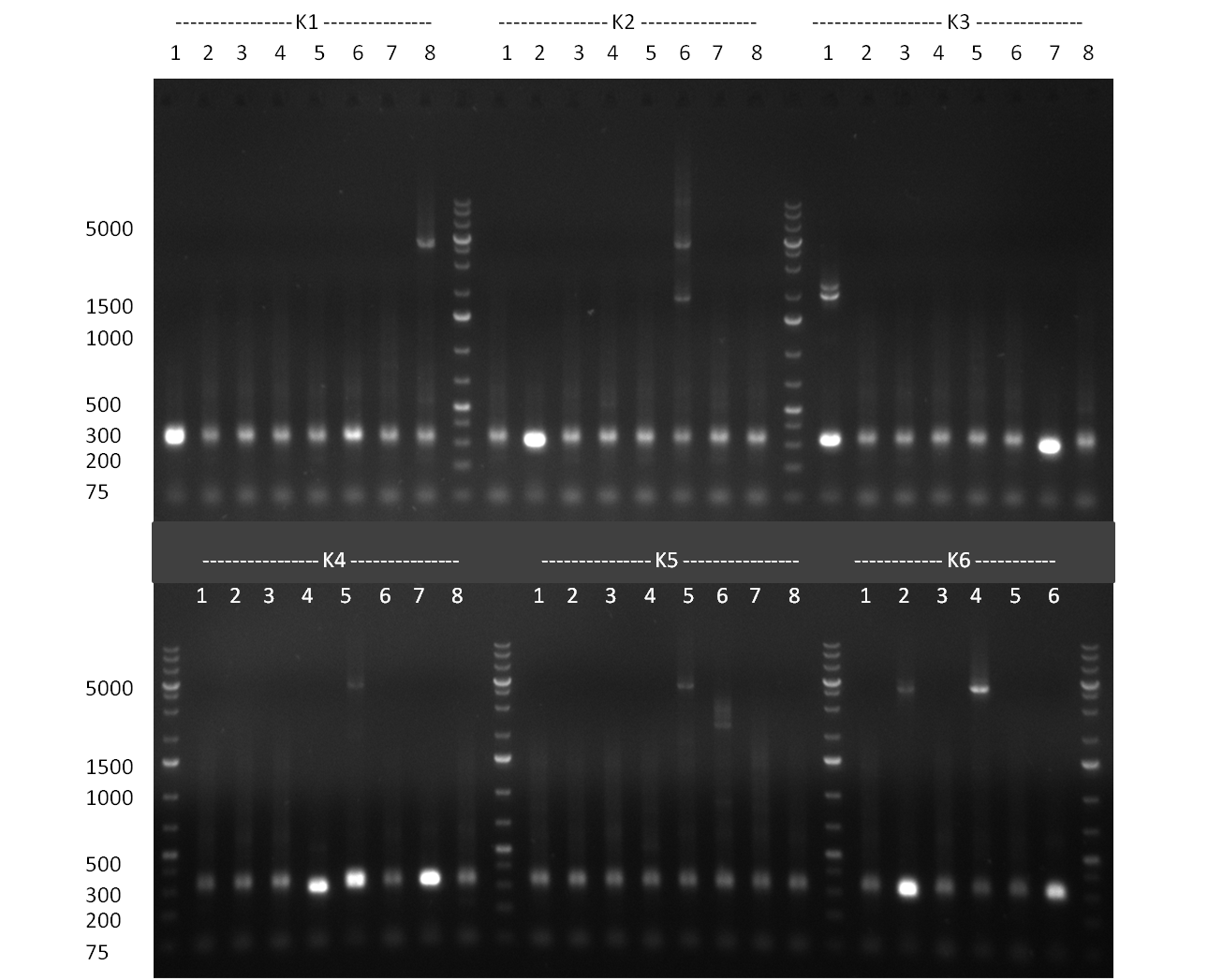
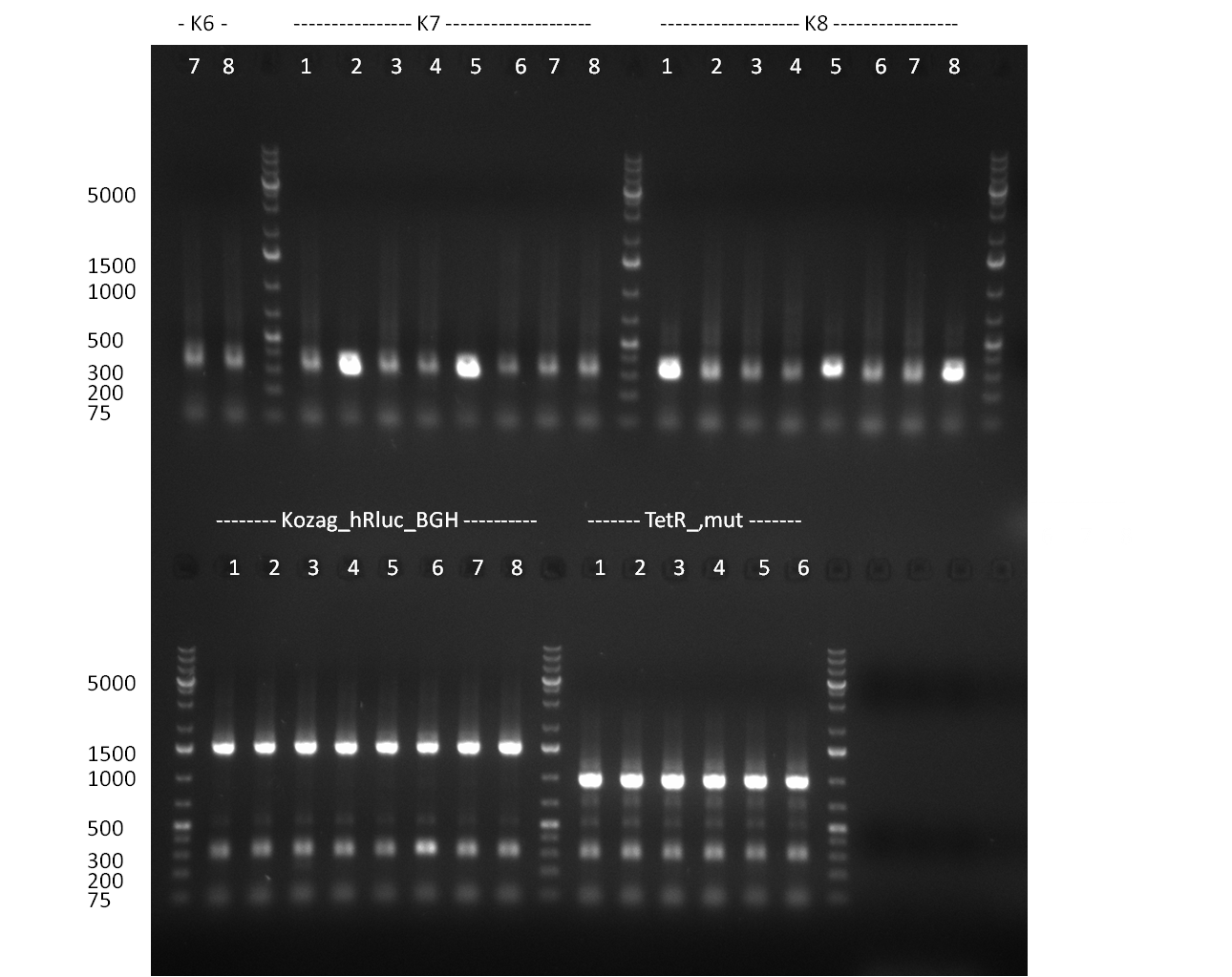
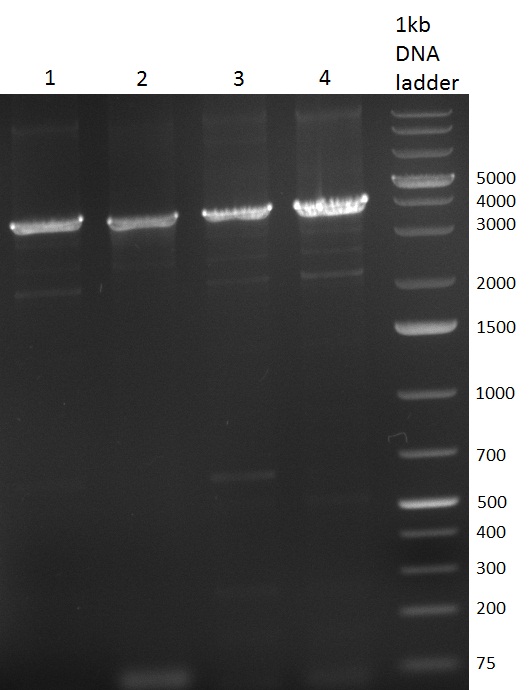
* Selection of colonies from the plates (see previous days' cloning) via colony-PCR using the standard sequencing primers; a ~5.5 kb fragment should be visible for the assembled construct K1-K8, if colony-PCR was positive (gel 100917-1 and 100917-2). No sample shows a 5.5 kb band, but that's most likely due to the very difficult amplification of such a long fragment with Fermentas 2x PCR MasterMix (Taq-based). Therefor 3 miniprep cultures were inocculated for each construct (K1-K8). The TetR oligo insertion and the Kozag_hRluc_BGH cloning into pSB1C3 gave the right bands on the gel. | * Selection of colonies from the plates (see previous days' cloning) via colony-PCR using the standard sequencing primers; a ~5.5 kb fragment should be visible for the assembled construct K1-K8, if colony-PCR was positive (gel 100917-1 and 100917-2). No sample shows a 5.5 kb band, but that's most likely due to the very difficult amplification of such a long fragment with Fermentas 2x PCR MasterMix (Taq-based). Therefor 3 miniprep cultures were inocculated for each construct (K1-K8). The TetR oligo insertion and the Kozag_hRluc_BGH cloning into pSB1C3 gave the right bands on the gel. | ||
| - | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | |
===19/09/2010=== | ===19/09/2010=== | ||
| Line 289: | Line 356: | ||
<br /> | <br /> | ||
* test digestion of the miniprepped clones (previous day) with EcoRI/PstI; For all clones at least 2 digestions gave the exactly expected band (gel 100920-1 and 100920-2); therefor we can say, that the tuning construct is succesfully assembled and ready to be tested | * test digestion of the miniprepped clones (previous day) with EcoRI/PstI; For all clones at least 2 digestions gave the exactly expected band (gel 100920-1 and 100920-2); therefor we can say, that the tuning construct is succesfully assembled and ready to be tested | ||
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
| + | |||
| + | |||
| - | |||
| - | |||
===22/09/2010=== | ===22/09/2010=== | ||
| - | * digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] with EcoRI and PstI in EcoRI Buffer (NEB) and ligation into digested | + | * digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] with EcoRI and PstI in EcoRI Buffer ([http://www.neb.com NEB]) and ligation into digested [http://partsregistry.org/Part:pSB1C3 pSB1C3], transformation, growing on selective agar plates (chloramphenicol), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] |
| - | + | [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay]<br/> | |
| + | In order to test for promoter efficiency and to check whether the miRNA kit assembly works fine | ||
| + | 50ng of each construct with different promoter set-ups were transfected into HEK 293 T-REx cells and other cell lines HEK, HeLa, Huh7 in 96-well plate format using FuGENE transfection reagent. As every construct is expressing firefly luciferase (luc2) and renilla luciferase (hRluc) at the same time the setup allows for standardization of transfection efficiency as only luc2 is tagged with binding sites. Each sample was transfected and measured by Dual luciferase assay in 8 replicates. As by this time no shRNA has been cloned into plasmid no knock-down of luc2 is expected and the different expression efficiencies allow for characterization of the different promoters. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 BBa_K337032] leads to a relative luciferase unit (RLU) of luc2 to hRluc expression of 6 RLU. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337035 BBa_K337035] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337035 BBa_K337035] are showing a comparable expression of 12 - 13 RLU, which is in line with the knowledge that both luciferases are driven by the CMV promoter. Hek 293 T-Rex cells stably express the Tet repressor thus allows us to observe very efficient repression of Firefly luciferse expression if a CMV-TetO2 promoter is driving luc2 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 BBa_K337038] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337046 BBa_K337046]). [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337040 BBa_K337040] transfection into Hek T-Rex cells results in an expression of 15 RLU. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337042 BBa_K337042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337044 BBa_K337044] are constructed in a way that luc2 is driven by the CMV promoter and hRluc is driven by the RSV promoter and show a comparable expression of 17-20 RLU. This leads to the conclusion that the CMV promoter shows comparable expression to the RSV promoter in Hek T-Rex cell lines. If transfecting [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337035 BBa_K337035] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 BBa_K337036] are transfected into different cell lines it is obvious that Hek293T cells are the easiest to transfect with both constructs an expression of 17-22 RLU is to be measured. Hek T-Rex cells are showing and expression level of 12 RLU of both constructs. Hela cells are also showing constant expression levels of 8 RLU with both constructs. A rather high standard deviation in the luciferase expression and also differences between the 2 constructs is to be seen in Huh7 cells. This might be due to low transfection efficiency of this cell line in general. Alltogether it is to say that [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337035 BBa_K337035] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 BBa_K337036] show comparable expression. | ||
| + | |||
| + | |||
| + | {| class="wikitable sortable" border="0" style="text-align: center" | ||
| + | |-bgcolor=#cccccc | ||
| + | !part!!promoter driving luc2 (Firefly)!!promoter driving (Renilla)!!promoter driving shRNA expression | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 BBa_K337032]||RSV||CMV||SV40 | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337035 BBa_K337035]||CMV||CMV||SV40 | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 BBa_K337036]||CMV||CMV||RSV | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 BBa_K337038]||CMV TetO2||CMV ||RSV | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337040 BBa_K337040]||RSV||RSV||SV40 | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337042 BBa_K337042]||CMV||RSV||SV40 | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337044 BBa_K337044]||CMV||RSV||RSV | ||
| + | |- | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K337046 BBa_K337046]||CMV TetO2||RSV||RSV | ||
| + | |- | ||
| + | |} | ||
| + | <br /><br /><br /> | ||
| + | [[Image:Promoter_test_220910_hd2010.jpg| thumb | 600px | centre | Promoter strength characterization in HEK 293 T-REx cell line]] | ||
| + | <br /><br /><br /> | ||
| + | [[Image:23092010 DLA promoters cell lines.jpg | thumb | 600px | centre| Promoter strength characterization in different cell lines]] | ||
| + | |||
| + | <br /> | ||
===23/09/2010=== | ===23/09/2010=== | ||
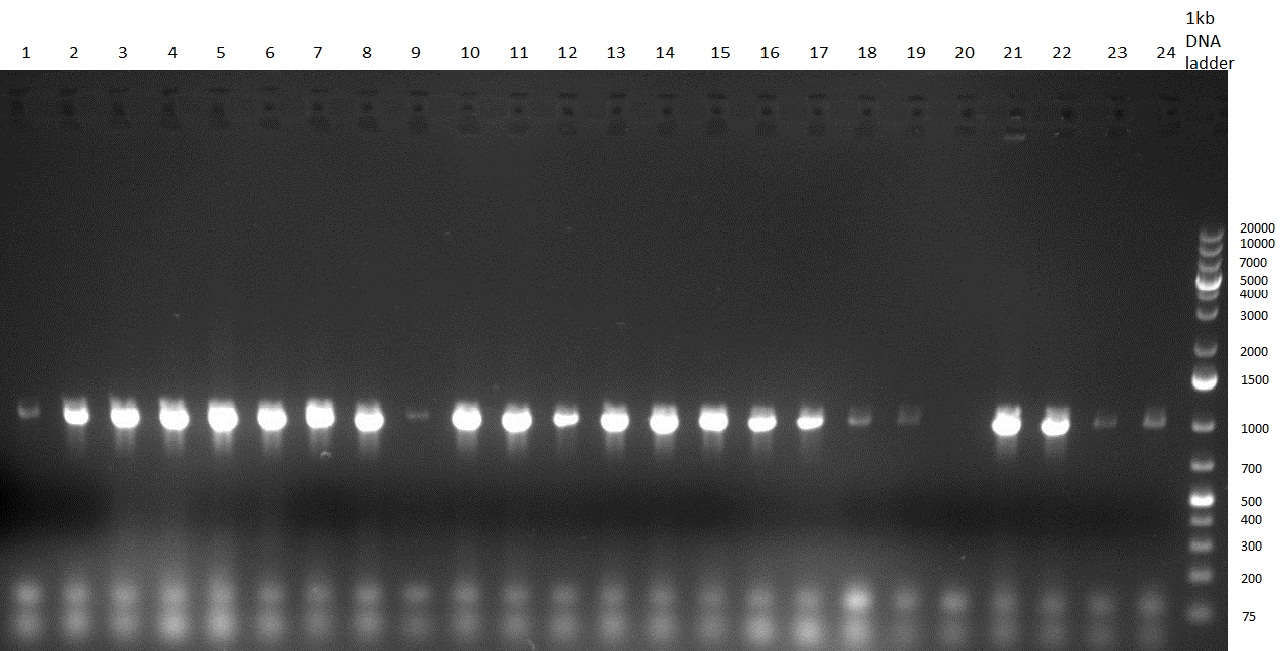
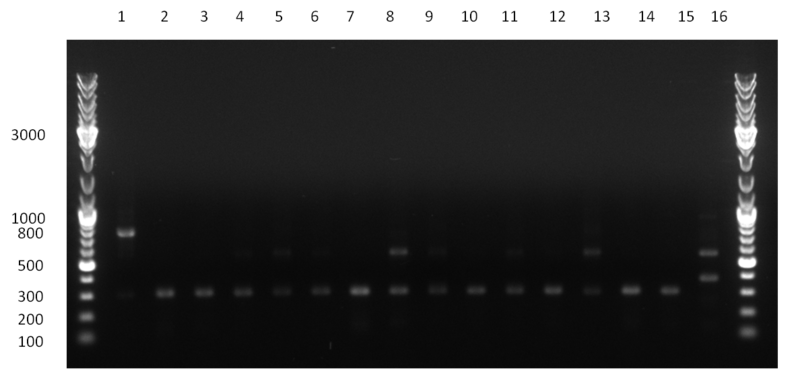
[[Image:20100923 LoLa colonypcr.png | thumb | 350px | right | Gel 100923-1. Analytical gel with following lanes: 1) till 8) colony PCR products with nice bands as expected at <nowiki>~</nowiki> 800 bp especially at lane 1, 4, 6 and 8, lane 9) 1kb Plus Ladder (Invitrogen)]] | [[Image:20100923 LoLa colonypcr.png | thumb | 350px | right | Gel 100923-1. Analytical gel with following lanes: 1) till 8) colony PCR products with nice bands as expected at <nowiki>~</nowiki> 800 bp especially at lane 1, 4, 6 and 8, lane 9) 1kb Plus Ladder (Invitrogen)]] | ||
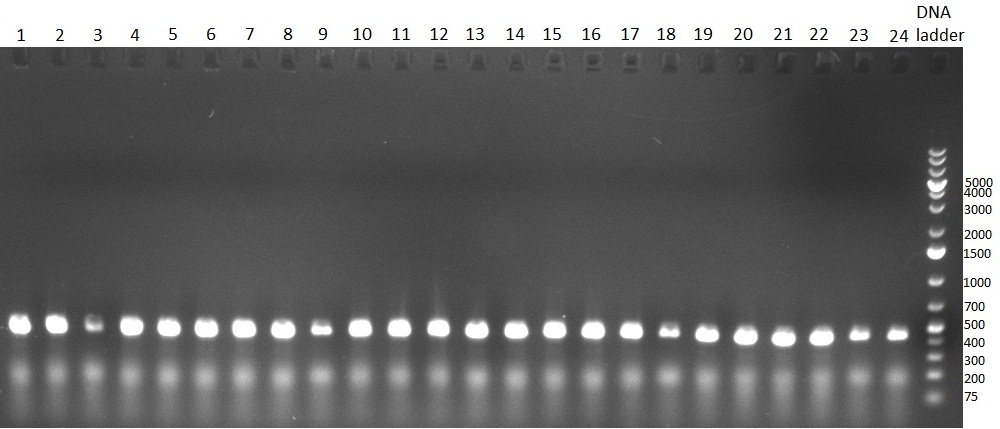
| - | * minipreps (using Quiagen Kit) and [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of each 8 colonies of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] in | + | * minipreps (using Quiagen Kit) and [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] of each 8 colonies of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] in [http://partsregistry.org/Part:pSB1C3 pSB1C3] (digested, ligated, transformed and plated on [[Igem2010/Main/synthetic_miR_Kit/September#22/09/2010 | previous day]]) |
*** PCR product of R23C on analytical gel (see gel 100923-1) | *** PCR product of R23C on analytical gel (see gel 100923-1) | ||
** digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R33]] with either SpeI and PstI or EcoRI and NheI | ** digestion of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R23]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R33]] with either SpeI and PstI or EcoRI and NheI | ||
| - | ** ligation into digested | + | ** ligation into digested [http://partsregistry.org/Part:pSB1A3 pSB1A3], [https://2010.igem.org/Transformation transformation], growing on selective agar plates (ampicillin), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] |
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /> |
| + | [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay] was performed as on [[https://2010.igem.org/Team:Heidelberg/Notebook/miRNA_Kit/September#22.2F09.2F2010 previous day]]. Cells were not harvested after 20h, but 48h. | ||
| + | |||
| + | <br /><br /><br /><br /><br /><br /> | ||
===24/09/2010=== | ===24/09/2010=== | ||
| - | |||
*inoculation and colony PCR of the transformed (23A+33A)=56C construct from yesterday. Same PCR protocol was used, even though the fragment should be 1100 and therefore elongation was probably not long enough. The PCR showed a band at 300bp, which dominik says is normal if you have religated vector. But nevertheless, miniprep of La1, La3 and La4 were made, because they didn't show a strong negative PCR band. | *inoculation and colony PCR of the transformed (23A+33A)=56C construct from yesterday. Same PCR protocol was used, even though the fragment should be 1100 and therefore elongation was probably not long enough. The PCR showed a band at 300bp, which dominik says is normal if you have religated vector. But nevertheless, miniprep of La1, La3 and La4 were made, because they didn't show a strong negative PCR band. | ||
| Line 320: | Line 420: | ||
* ligation of R56C + R8 + backbone A | * ligation of R56C + R8 + backbone A | ||
<br> | <br> | ||
| + | |||
===25/09/2010=== | ===25/09/2010=== | ||
* PCR amplification of standardised pcDNA5 p55 and p1 with primer D47/48 and D55/69 | * PCR amplification of standardised pcDNA5 p55 and p1 with primer D47/48 and D55/69 | ||
| + | <br /> | ||
===26/09/2010=== | ===26/09/2010=== | ||
[[Image:20100926 digestion check.png | thumb | 350px | right | Gel 100926-1. Analytical gel with following lanes: 1) R33 E/N (from 23<sup>rd</sup> September) 2) R23_1 S/P 3) R23_6 4) R33C 5) R23_4 6) R23_8 7) 1kb Plus Ladder (Invitrogen) 8) R33C E/N 9) R23_4 S/P 9) R23C_8 S/P]] | [[Image:20100926 digestion check.png | thumb | 350px | right | Gel 100926-1. Analytical gel with following lanes: 1) R33 E/N (from 23<sup>rd</sup> September) 2) R23_1 S/P 3) R23_6 4) R33C 5) R23_4 6) R23_8 7) 1kb Plus Ladder (Invitrogen) 8) R33C E/N 9) R23_4 S/P 9) R23C_8 S/P]] | ||
| - | [[Image:Rb100927_pcrp55andp1.jpg|thumb| | + | [[Image:Rb100927_pcrp55andp1.jpg|thumb|300px|right| Gel 100926-2. PCR results of p55 and p1: p1 with primer 47/48 (lane 1), p1 with primer 55/70 (lane 2), p55 with primer 47/48 (lane3), p55 with primer 55/70]] |
* miniprep of R23C cultures again (colonies from transformation (done [[Igem2010/Main/synthetic miR Kit/September#22/09/2010 | on wednesday]])) | * miniprep of R23C cultures again (colonies from transformation (done [[Igem2010/Main/synthetic miR Kit/September#22/09/2010 | on wednesday]])) | ||
* check of all digestions on an analytical gel (see gel 100926-1) | * check of all digestions on an analytical gel (see gel 100926-1) | ||
| Line 332: | Line 434: | ||
**** because of reverse elements and resulting secondary structure? | **** because of reverse elements and resulting secondary structure? | ||
** repetition of ligation with R23_6 and old R33 (nice band at roughly 600bp) | ** repetition of ligation with R23_6 and old R33 (nice band at roughly 600bp) | ||
| - | *** including negative control at this time with only backbone | + | *** including negative control at this time with only backbone [http://partsregistry.org/Part:pSB1A3 pSB1A3] |
** [https://2010.igem.org/Transformation transformation] | ** [https://2010.igem.org/Transformation transformation] | ||
<br> tuning construct: | <br> tuning construct: | ||
* gel extraction of PCR on [[Igem2010/Main/synthetic miR Kit/September#25/09/2010 | p55 and p1]] (see gel 100926-2) | * gel extraction of PCR on [[Igem2010/Main/synthetic miR Kit/September#25/09/2010 | p55 and p1]] (see gel 100926-2) | ||
| - | * recloning of R1,R4 and R7 into standardbackbone | + | * recloning of R1,R4 and R7 into standardbackbone [http://partsregistry.org/Part:pSB1A3 pSB1A3] |
** digest 500ng of R1, R4 and R7 with EcoRI and PstI | ** digest 500ng of R1, R4 and R7 with EcoRI and PstI | ||
** heat inactivate 20 min at 80°C | ** heat inactivate 20 min at 80°C | ||
** Nucleotide removal | ** Nucleotide removal | ||
| - | ** ligate 150ng and 250ng of insert into 30ng | + | ** ligate 150ng and 250ng of insert into 30ng [http://partsregistry.org/Part:pSB1A3 pSB1A3] 1h at RT and transform ligation |
| - | <br><br><br><br> | + | <br><br><br><br><br><br> |
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
===27/09/2010=== | ===27/09/2010=== | ||
| Line 349: | Line 451: | ||
<br/> | <br/> | ||
tuning construct: | tuning construct: | ||
| - | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] on [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K1]], [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K4]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K7]] cloned into | + | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] on [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K1]], [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K4]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K7]] cloned into [http://partsregistry.org/Part:pSB1A3 pSB1A3] showed nearly only positives as a band of 400bp is expected by using the primer pair 50/75 |
* miniprep of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K1]], [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K4]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K7]] | * miniprep of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K1]], [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K4]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K7]] | ||
* send for sequencing | * send for sequencing | ||
| - | * cloning of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K3]] into | + | * cloning of [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K3]] into [http://partsregistry.org/Part:pSB1A3 pSB1A3]: |
** digest [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K3]] with PstI and EcoRI | ** digest [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | K3]] with PstI and EcoRI | ||
| - | ** ligate 200ng with 30ng of vector | + | ** ligate 200ng with 30ng of vector [http://partsregistry.org/Part:pSB1A3 pSB1A3] |
** transform ligation and plate on an Ampicillin plate | ** transform ligation and plate on an Ampicillin plate | ||
* digest shRNA 6 (F11) and shRNA 7 (F12): | * digest shRNA 6 (F11) and shRNA 7 (F12): | ||
| Line 365: | Line 467: | ||
* a lot of religations, thus: a lot of colonies to pick | * a lot of religations, thus: a lot of colonies to pick | ||
* [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] and test digestion with SpeI and PstI of promising mini-preps revealed some positive samples | * [[Igem2010/Main/Protocols/Colony_PCR | colony-PCR]] and test digestion with SpeI and PstI of promising mini-preps revealed some positive samples | ||
| - | * ligation with [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R8]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R11]], respectively (each digested with EcoRI and NheI) in | + | * ligation with [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R8]] and [[Igem2010/Main/Library/Constructs/pSMB miTrigger#basal components and cloning strategy | R11]], respectively (each digested with EcoRI and NheI) in [http://partsregistry.org/Part:pSB1C3 pSB1C3] (backbone, digested with EcoRI and PstI), following [https://2010.igem.org/3A_Assembly standard protocol recommendations] |
* growth of transformed cells on selective agar plates (chloramphenicol) over night | * growth of transformed cells on selective agar plates (chloramphenicol) over night | ||
| - | + | <br><br><br> | |
===28/09/2010=== | ===28/09/2010=== | ||
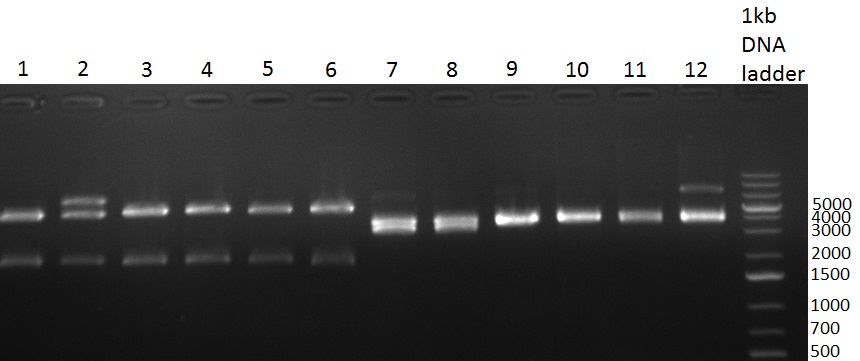
| - | [[Image:Rb100928_K3.jpg|thumb|350px| | + | [[Image:Rb100928_K3.jpg|thumb|350px|right| Gel 100928-1. Colony PCR of K3 construct]] |
[[Image:Rb100928_K3andtestdigestK147.jpg|thumb|350px|right| Gel 100928-2. Test digest of with PstI and EcoRI (lane 1-6), test digest with BamHI (lane 7-12), K1 (lane 1&2), K4 (lane 3&4), K7 (lane 5&6)]] | [[Image:Rb100928_K3andtestdigestK147.jpg|thumb|350px|right| Gel 100928-2. Test digest of with PstI and EcoRI (lane 1-6), test digest with BamHI (lane 7-12), K1 (lane 1&2), K4 (lane 3&4), K7 (lane 5&6)]] | ||
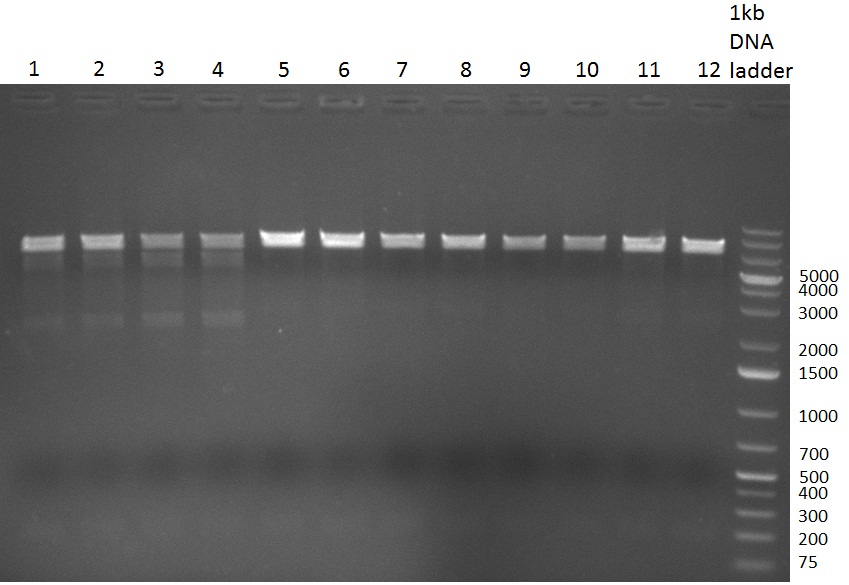
| - | [[Image:Rb100928_digestk147ah.jpg|thumb|350px| | + | [[Image:Rb100928_digestk147ah.jpg|thumb|350px|right| Gel 100928-3. Digestion of K1 (lane 1-4), K4 (lane 5-8) and K7 (lane 9-12) with HindIII and AflII]] |
tuning construct: | tuning construct: | ||
* colony PCR of K3 (gel 100928-1) | * colony PCR of K3 (gel 100928-1) | ||
| Line 392: | Line 494: | ||
<br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br> | ||
| - | <br><br><br><br><br><br><br><br><br><br> | + | <br><br><br><br><br><br><br><br><br><br><br /><br /><br /><br /> |
| - | <br><br><br><br> | + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
| - | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> | + | |
===29/09/2010=== | ===29/09/2010=== | ||
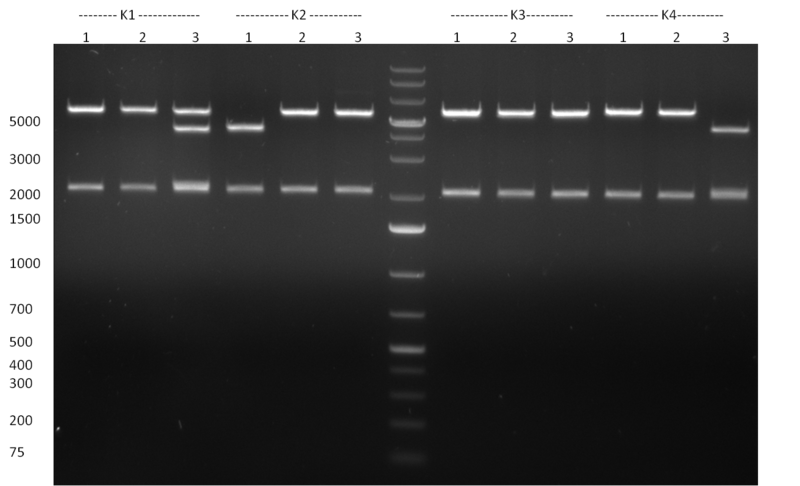
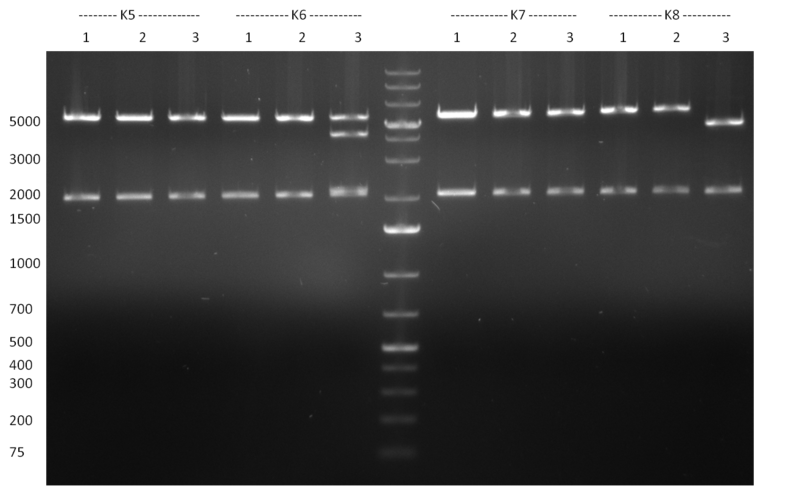
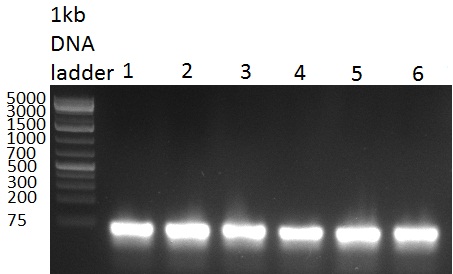
| - | [[Image:Rb100928testpe_bk1k3.jpg|thumb|350px| | + | [[Image:Rb100928testpe_bk1k3.jpg|thumb|350px|right|Gel 100929-1 first gel: double digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with PstI and EcoRI: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1-4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] (lane 5-8), second gel: digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with BamHI: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1-4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] (lane 5-8)]] |
| - | [[Image:Rb100929k1k3ahdigest.jpg|thumb|350px|right|Gel 1000929-2 digestion of K1 and K3 with HindIII and AflII: K1 (lane 1&2), K3(lane 3-12)]] | + | [[Image:Rb100929k1k3ahdigest.jpg|thumb|350px|right|Gel 1000929-2 digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with HindIII and AflII: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1&2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3](lane 3-12)]] |
[[Image:20100929_gel1_bs_ligationcontrol.jpg|thumb|350px|right|gel 100929-3 pcr on binding sites for shRNA 10]] | [[Image:20100929_gel1_bs_ligationcontrol.jpg|thumb|350px|right|gel 100929-3 pcr on binding sites for shRNA 10]] | ||
tuning construct: | tuning construct: | ||
| - | * mini Prep on K3 and K1 constructs | + | * mini Prep on [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] constructs |
** test digestion with PstI & EcoRI and BamHI showed all positive results | ** test digestion with PstI & EcoRI and BamHI showed all positive results | ||
| - | ** digest K3 and K1 with HindIII and AflII | + | ** digest [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] with HindIII and AflII |
| - | * cloning of shRNA 6 and 7 into K1, K3, K4 and K7: | + | * cloning of shRNA 6 and 7 into [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337044 K7]: |
** colony PCR on ligation from the 28/09/2010 did not reveal any positives by using the following protocol: | ** colony PCR on ligation from the 28/09/2010 did not reveal any positives by using the following protocol: | ||
** colony PCR on ligations from the 28/09/2010 did not reveal any positives by using the following protocol: | ** colony PCR on ligations from the 28/09/2010 did not reveal any positives by using the following protocol: | ||
** start from the beginning again: in doublicates: | ** start from the beginning again: in doublicates: | ||
** first procedure: | ** first procedure: | ||
| - | *** digest PCR on shRNA 6 and 7 as well as K1, K3, K4 and K7 with HindIII and AflII | + | *** digest PCR on shRNA 6 and 7 as well as [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337044 K7] with HindIII and AflII |
*** purify shRNAs with nucleotide removal kit | *** purify shRNAs with nucleotide removal kit | ||
| - | *** purify digested K3 and K1 with gel extraction kit | + | *** purify digested [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] with gel extraction kit |
| - | *** ligate K1, K3, K4 and K7 with shRNA 6 and 7 | + | *** ligate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337044 K7] with shRNA 6 and 7 |
** second procedure: | ** second procedure: | ||
| - | *** digest K4 and K1 as well as shRNA6 and 7 from PCR product and purify via nucleotide removal kit | + | *** digest [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] as well as shRNA6 and 7 from PCR product and purify via nucleotide removal kit |
| - | *** ligate K4 and K3 with shRNA 6 and 7 | + | *** ligate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with shRNA 6 and 7 |
** transform all ligations | ** transform all ligations | ||
* cloning of binding sites: | * cloning of binding sites: | ||
| Line 429: | Line 530: | ||
* R8 and R11 were digested with E/N again | * R8 and R11 were digested with E/N again | ||
* digestion of P56 colony 14 was positive on the gel, digestion of R8 and R11 also gave the right fragments | * digestion of P56 colony 14 was positive on the gel, digestion of R8 and R11 also gave the right fragments | ||
| - | * | + | * [http://partsregistry.org/Part:pSB1C3 pSB1C3] backbone was [http://www.fermentas.com/templates/files/tiny_mce/coa_pdf/coa_ef0511.pdf SAP] treated to reduce religation that was likely to be the problem with previous transformations |
* ligation of P56 (14) with R8 and R11 and SAPed backbone and control ligation with backbone only, transformation | * ligation of P56 (14) with R8 and R11 and SAPed backbone and control ligation with backbone only, transformation | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| - | <br/><br/><br/><br/><br/><br/> | + | <br/><br/><br/><br/><br/><br/><br /><br /><br /> |
| - | <br/><br/><br/><br/><br/><br/> | + | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
<br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| - | |||
===30/09/2010=== | ===30/09/2010=== | ||
| Line 441: | Line 541: | ||
[[Image:20100930_gel2.jpg|thumb|350px|right|Gel 1000930-3 colony PCR on shRNA10 into K3 and K4]] | [[Image:20100930_gel2.jpg|thumb|350px|right|Gel 1000930-3 colony PCR on shRNA10 into K3 and K4]] | ||
tuning construct: | tuning construct: | ||
| - | * colony pcr on shRNA 6 and 7 into construct K3 and K4 | + | * colony pcr on shRNA 6 and 7 into construct [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] |
| - | * cloning of binding sites for shRNA 10 into K3 and K4: | + | * cloning of binding sites for shRNA 10 into [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4]: |
| - | ** digestion of binding sites (perfect, imperfect 9-12 and imperfect 9-22) for shRNA10 with xhoI and ageI :) and construct K3 and K4 in | + | ** digestion of binding sites (perfect, imperfect 9-12 and imperfect 9-22) for shRNA10 with xhoI and ageI :) and construct [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337038 K4] in [http://partsregistry.org/Part:pSB1A3 pSB1A3] with xmaI and xhoI |
** ligation of binding sites with digested constructs | ** ligation of binding sites with digested constructs | ||
** transformation of ligation into TOP10 cells | ** transformation of ligation into TOP10 cells | ||
** plate on ampicillin plates ON | ** plate on ampicillin plates ON | ||
| - | [[Image:20100930 lola colonypcr positiv!!!!!anno.jpg|thumb|350px| | + | [[Image:20100930 lola colonypcr positiv!!!!!anno.jpg|thumb|350px|right|Gel 100930-1 colony PCR of Repressor construct with different promoters (R8 or R11). Positive colonies show an amplification product at 2500bp (lanes 1,2,4-8, 10, 13-16)]] |
* plenty of colonies on the P56+R8 and P56+R11, NO colonies on control plate with SAPed backbone, colony PCR revealed 50% of positive clones | * plenty of colonies on the P56+R8 and P56+R11, NO colonies on control plate with SAPed backbone, colony PCR revealed 50% of positive clones | ||
<br/> | <br/> | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 05:57, 27 October 2010

01/09/2010
02/09/2010
03/09/2010
04/09/2010
05/09/2010
06/09/2010
07/09/2010
All Ligations were controlled by loading 10 ul of ligation reactiong onto a 1 % agarose gel (100907-1), run for 35 min @ 100 V
08/09/2010
09/09/2010
10/09/2010
Subsequent Transformation into DH5alpha cells
11/09/2010
12/09/2010
13/09/2010
14/09/2010
15/09/2010
Each PCR was performed in two replicates. The result of the PCRs was analyzed on a 1 % agarose gel (100915-1), run for 1 h @ 100 V. The PCR nr. 2 was subsequently PCR purified by applying a Qiagen PCR purification KIT and used for the cloning 16/09/2010
17/09/2010
19/09/2010
20/09/2010
22/09/2010
23/09/2010
24/09/2010
25/09/2010
26/09/2010
27/09/2010
28/09/2010tuning construct:
29/09/2010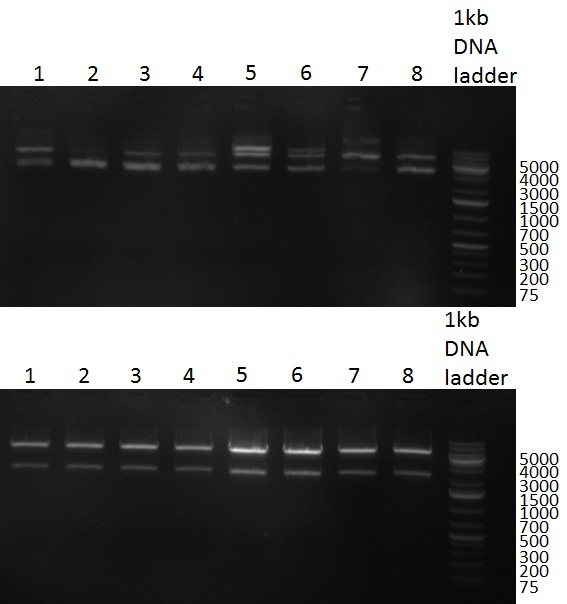 Gel 100929-1 first gel: double digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with PstI and EcoRI: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1-4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] (lane 5-8), second gel: digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with BamHI: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1-4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] (lane 5-8)  Gel 1000929-2 digestion of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3] with HindIII and AflII: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337032 K1] (lane 1&2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K337036 K3](lane 3-12) tuning construct:
 Gel 100929 r-5: test digestion of miniprep number 14 withe EcoRI and NheI shows expected band at 1100bp. Digestions of R8 and R11 were also positive, DNA cut out with SpeI and PstI runs at 800bp. Lane 1=miniprep 14 undigested, lane2=miniprep 14 digested, lane3=miniprep 23 digested, lane4=R8 undigested, lane5= R8 digested, lane6=R11 undigested, lane7=R11 digested repressor construct:
30/09/2010tuning construct:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"