Project/SpooA/
From 2010.igem.org
(→BioBrick Background) |
(→References) |
||
| (6 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
=== BioBrick Background === | === BioBrick Background === | ||
| - | So as to inhibit the transcription driven by the PlcR promoter, the SpoOA protein binds to a particular sequence | + | So as to inhibit the transcription driven by the PlcR promoter, the SpoOA protein binds to a particular sequence. Binding to this region prevents polymerase from properly interacting with the promoter and thus represses transcription. The regions within the PlcR promoter that SpoOA binds are referred to as the SpoOA boxes, of which, two of these boxes flank the PlcR-PapR positive regulator binding site thus repressing transcription [1]. |
| + | |||
| + | === Sequence Development and Annotation === | ||
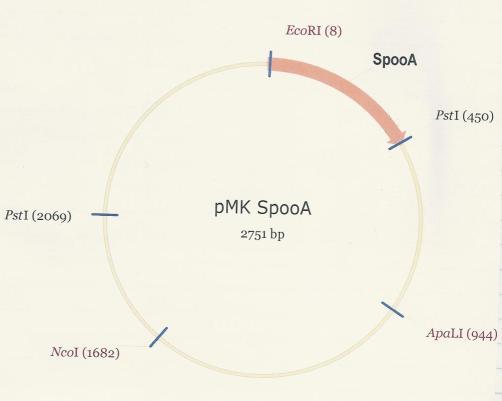
| + | The SpoOA repressor was synthesized and and delivered within the pMK plasmid. So as to use SpoOA in development, it was excised from the plasmid via digestion with EcoR1 and Pst1. <br /> | ||
| + | [[Image:PMK-SpoOA_-real.JPG]] <br /> <br /> | ||
| + | |||
| + | Once digested, the SpoOA fragment was gel extracted and purified in preparation for ligation into PSB1C3. The SpoOA insert (442bp) can be clearly seen. <br /> | ||
| + | [[Image:SpoOA_digest.JPG]] <br /> <br /> | ||
| + | |||
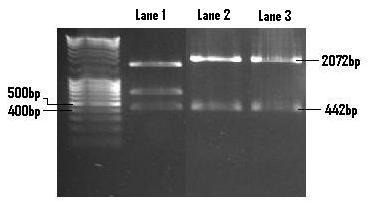
| + | The retrieved SpoOA fragments were ligated into the PSB1C3 linearised plasmid and then re-digested with EcoR1 and Pst1 so as to confirm the insertion. The SpoOA fragment (442bp) as well as the PSB1C3 linearised plasmid (2072bp) are both clearly visible in Lane 3. Lane 1 contains the pMK-SpoOA digested control with corresponding banding pattern. <br /> | ||
| + | [[Image:SpoOA_confirmation_digest.JPG]] <br /> <br /> | ||
| + | |||
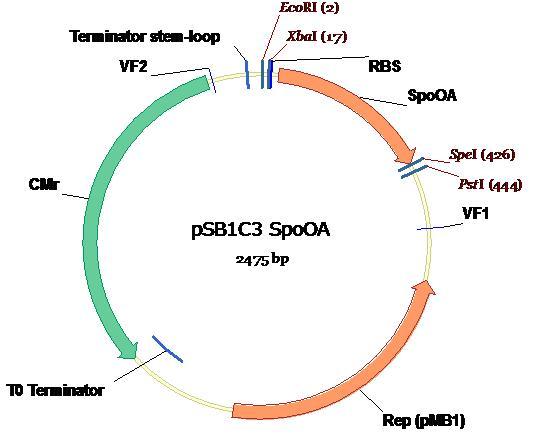
| + | The SpoOA-PSB1C3 plasmid map is shown below as well as all necessary restriction sites and landmark annotations [2].<br /> | ||
| + | [[Image:PMK-SpoOA.JPG]] <br /> | ||
| - | |||
| - | |||
=== Testing and verification === | === Testing and verification === | ||
<br /> | <br /> | ||
| + | === References === | ||
| + | [1] Lereclus D., Agaisse H., Grandvalet C., Salamitou S. & Gominet M. (2000), Regulation of toxin and virulence gene transcription in ''Bacillus thuringiensis''. Int. J. Med. Microbiol. '''290:'''295-299. <br /> | ||
| + | [2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za. | ||
Latest revision as of 20:32, 25 October 2010

Contents |
SpooA repressor - [http://partsregistry.org/Part:BBa_K354004 BBa_K354004]
BioBrick Background
So as to inhibit the transcription driven by the PlcR promoter, the SpoOA protein binds to a particular sequence. Binding to this region prevents polymerase from properly interacting with the promoter and thus represses transcription. The regions within the PlcR promoter that SpoOA binds are referred to as the SpoOA boxes, of which, two of these boxes flank the PlcR-PapR positive regulator binding site thus repressing transcription [1].
Sequence Development and Annotation
The SpoOA repressor was synthesized and and delivered within the pMK plasmid. So as to use SpoOA in development, it was excised from the plasmid via digestion with EcoR1 and Pst1.
Once digested, the SpoOA fragment was gel extracted and purified in preparation for ligation into PSB1C3. The SpoOA insert (442bp) can be clearly seen.

The retrieved SpoOA fragments were ligated into the PSB1C3 linearised plasmid and then re-digested with EcoR1 and Pst1 so as to confirm the insertion. The SpoOA fragment (442bp) as well as the PSB1C3 linearised plasmid (2072bp) are both clearly visible in Lane 3. Lane 1 contains the pMK-SpoOA digested control with corresponding banding pattern.
The SpoOA-PSB1C3 plasmid map is shown below as well as all necessary restriction sites and landmark annotations [2].
Testing and verification
References
[1] Lereclus D., Agaisse H., Grandvalet C., Salamitou S. & Gominet M. (2000), Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int. J. Med. Microbiol. 290:295-299.
[2] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za.
 "
"