Team:Lethbridge/Notebook/Protocols
From 2010.igem.org
Adam.smith4 (Talk | contribs) (→Maxiprep) |
Liszabruder (Talk | contribs) |
||
| (13 intermediate revisions not shown) | |||
| Line 16: | Line 16: | ||
<th> | <th> | ||
| - | <image src="https://static.igem.org/mediawiki/2010/ | + | <image src="https://static.igem.org/mediawiki/2010/9/91/UofLLabWork.JPG" height="300px"/> |
</th> | </th> | ||
| Line 147: | Line 147: | ||
==<font color="white">Assembly of BioBricks using the <font color="red">R</font>e<font color="red">d</font>/<font color="red">W</font>h<font color="red">i</font>t<font color="red">e</font> 3-Antibiotic Assembly Method== | ==<font color="white">Assembly of BioBricks using the <font color="red">R</font>e<font color="red">d</font>/<font color="red">W</font>h<font color="red">i</font>t<font color="red">e</font> 3-Antibiotic Assembly Method== | ||
| + | |||
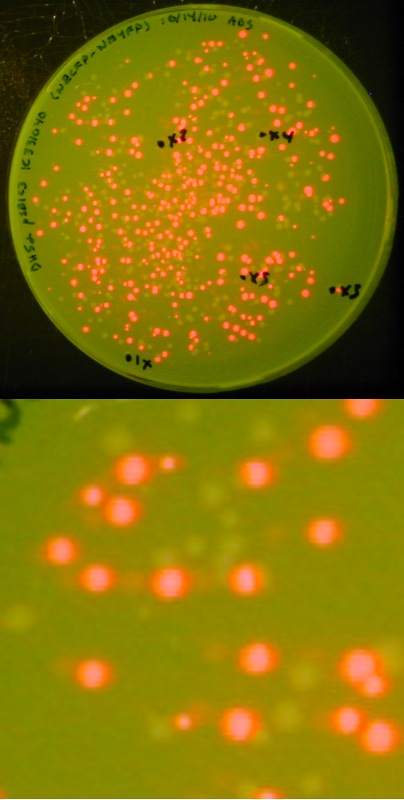
[[image:Lethbridge 101021RFP.jpg|200px|right|Easy visual screening of transformed <i>Escherichia coli</i> cells using the Red/White 3-antibiotic assembly method]] | [[image:Lethbridge 101021RFP.jpg|200px|right|Easy visual screening of transformed <i>Escherichia coli</i> cells using the Red/White 3-antibiotic assembly method]] | ||
This method is a variant of the 3-Antibiotic Assembly Method that has been developed by Ginkgo Bioworks and New England Biolabs.<br> | This method is a variant of the 3-Antibiotic Assembly Method that has been developed by Ginkgo Bioworks and New England Biolabs.<br> | ||
| - | *In the 3-Antibiotic Assembly Method, the destination backbone (< | + | *In the 3-Antibiotic Assembly Method, the destination backbone (<html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1A3" target="new"><font color="#00DC00"> pSB1A3</font></a></html>, <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1C3" target="new"><font color="#00DC00"> pSB1C3</font></a></html>, <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1K3" target="new"><font color="#00DC00"> pSB1K3</font></a></html>, or <html><a href="http://partsregistry.org/wiki/index.php/Part:pSB1T3" target="new"><font color="#00DC00"> pSB1T3</font></a></html>) is amplified via PCR, using DNA received in the 2010 Distribution. <br> |
We had difficulty generating a large quantity of plasmid backbone in this manner, and as a result were unable to assemble biobricks. <br><br> | We had difficulty generating a large quantity of plasmid backbone in this manner, and as a result were unable to assemble biobricks. <br><br> | ||
| - | As an alternative, we exploited the ability of part < | + | As an alternative, we exploited the ability of part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J04450" target="new"><font color="#00DC00"> BBa_J04450</font></a></html> (expressing red fluorescent protein - RFP) to create in its host cell a very strong red color following incubation; this has proven a useful and successful variant of the 3-Antibiotic Assembly Method.<br><br> |
We retained the selection advantage of the 3-Antibiotic Assembly Method (associated with having a destination plasmid containing a different antibiotic resistance than the upstream and downstream plasmid) as the expressing RFP biobrick is available in a wide variety of BBF plasmids. <br><br> | We retained the selection advantage of the 3-Antibiotic Assembly Method (associated with having a destination plasmid containing a different antibiotic resistance than the upstream and downstream plasmid) as the expressing RFP biobrick is available in a wide variety of BBF plasmids. <br><br> | ||
With this variation, we can not only visually screen our colonies for the presence of our expected assembly product (as indicated by a lack of red fluorescence), we also have a built-in negative control in that a lack of assembly product will result in a re-ligation of the expressing RFP biobrick into the destination backbone, producing red colonies.<br> | With this variation, we can not only visually screen our colonies for the presence of our expected assembly product (as indicated by a lack of red fluorescence), we also have a built-in negative control in that a lack of assembly product will result in a re-ligation of the expressing RFP biobrick into the destination backbone, producing red colonies.<br> | ||
| - | ===Step 1 - Restriction=== | + | |
| + | ===<font color="white">Step 1 - Restriction=== | ||
| + | |||
Performed according to Ginkgo Bioworks/NEB BioBrick Assembly Kit, with several modifications.<br> | Performed according to Ginkgo Bioworks/NEB BioBrick Assembly Kit, with several modifications.<br> | ||
<b>Digestion of upstream part</b> | <b>Digestion of upstream part</b> | ||
| Line 180: | Line 183: | ||
Following each restriction digest, all samples were subjected to heating at 80<sup>o</sup>C for 20 minutes to irreversibly denature the restriction endonucleases.<br><br> | Following each restriction digest, all samples were subjected to heating at 80<sup>o</sup>C for 20 minutes to irreversibly denature the restriction endonucleases.<br><br> | ||
† Typical concentrations obtained from our minipreps are approximately 50ng/µL, therefore adding 5µL of plasmid DNA gives the restriction reaction a concentration of 10ng/µL, as recommended in the NEB BioBrick Assembly Kit Literature. <br> | † Typical concentrations obtained from our minipreps are approximately 50ng/µL, therefore adding 5µL of plasmid DNA gives the restriction reaction a concentration of 10ng/µL, as recommended in the NEB BioBrick Assembly Kit Literature. <br> | ||
| - | ‡ Destination plasmid is either pSB1A3, pSB1C3, pSB1K3, or pSB1T3, containing part < | + | ‡ Destination plasmid is either pSB1A3, pSB1C3, pSB1K3, or pSB1T3, containing part <html><a href="http://partsregistry.org/wiki/index.php/Part:BBa_J04450" target="new"><font color="#00DC00">BBa_J04450</font></a></html> which is the expressing red fluorescent protein. |
| + | |||
| + | ===<font color="white">Step 2 - Ligation=== | ||
| - | |||
Performed according to Ginkgo Bioworks/NEB BioBrick Assembly Kit<br> | Performed according to Ginkgo Bioworks/NEB BioBrick Assembly Kit<br> | ||
<b>Ligation of Upstream and Downstream parts into Destination Plasmid</b><br> | <b>Ligation of Upstream and Downstream parts into Destination Plasmid</b><br> | ||
| Line 192: | Line 196: | ||
*11µL MilliQ H<sub>2</sub>O | *11µL MilliQ H<sub>2</sub>O | ||
Ligation mixes were incubated on the bench top (~20<sup>o</sup>C) for 10 minutes, then transformed into competent DH5α cells. | Ligation mixes were incubated on the bench top (~20<sup>o</sup>C) for 10 minutes, then transformed into competent DH5α cells. | ||
| - | ===Step 3 - Transformation=== | + | ===<font color="white">Step 3 - Transformation=== |
#Thaw 50µL of aliquotted competent cells (DH5α) on ice | #Thaw 50µL of aliquotted competent cells (DH5α) on ice | ||
#Gently pipet 2.0µL (~1ng) DNA (from ligation mix) into competent cells | #Gently pipet 2.0µL (~1ng) DNA (from ligation mix) into competent cells | ||
| Line 203: | Line 207: | ||
#Incubate the plates in the 37<sup>o</sup>C incubator for approximately 36 hours (agar on top) | #Incubate the plates in the 37<sup>o</sup>C incubator for approximately 36 hours (agar on top) | ||
| - | ===Step 4 - Selection of Colonies and Colony PCR=== | + | ===<font color="white">Step 4 - Selection of Colonies and Colony PCR=== |
| + | |||
Following ~36 hour incubation at 37<sup>o</sup>C, plates were inspected, and white colonies were picked and subsequently subjected to Colony PCR.<br> | Following ~36 hour incubation at 37<sup>o</sup>C, plates were inspected, and white colonies were picked and subsequently subjected to Colony PCR.<br> | ||
| - | <b>Colony PCR</b> | + | <b>Colony Picking</b> |
| + | #Pick a colony from a transformation plate using a sterile toothpick or micropipette tip | ||
| + | #Transfer the cells to a 1.5mL microcentrifuge tube containing 50µL of sterile MilliQ H<sub>2</sub>O | ||
| + | #Place microcentrifuge tube in heat block at 99<sup>o</sup>C for 5 minutes to lyse the cells and denature DNases. | ||
| + | #Centrifuge at max speed in a table-top microcentrifuge for 1 minute to remove cell debris | ||
| + | #Use 5µL of supernatant as template for PCR | ||
| + | <b>PCR to confirm length of newly assembled part</b><br> | ||
| + | Reaction Conditions: | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Concentration</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>N/A</td><td>33.8</td></tr> | ||
| + | <tr><td>Pfu Buffer with MgSO<sub>4</sub></td><td>10x</td><td>5</td></tr> | ||
| + | <tr><td>dNTP</td><td>10mM</td><td>2</td></tr> | ||
| + | <tr><td>VF2 Forward Primer</td><td>10µL</td><td>2</td></tr> | ||
| + | <tr><td>VR Reverse Primer</td><td>10µL</td><td>2</td></tr> | ||
| + | <tr><td>Template DNA</td><td>N/A</td><td>5</td></tr> | ||
| + | <tr><td>Pfu DNA Polymerase</td><td>2U/µL</td><td>0.2</td></tr></table> | ||
| + | Cycling Conditions: | ||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Step</b></td><td><b>Temperature</b><td><b>Time</b></td></tr> | ||
| + | <tr><td>1-Initial Denature</td><td>98<sup>o</sup>C</td><td>3 minutes</td></tr> | ||
| + | <tr><td>2-Denature</td><td>98<sup>o</sup>C</td><td>30 seconds</td></tr> | ||
| + | <tr><td>3-Anneal<sup>†</sup></td><td>58<sup>o</sup>C</td><td>30 seconds</td></tr> | ||
| + | <tr><td>4-Extend</td><td>72<sup>o</sup>C</td><td>30 seconds<sup>‡</sup></td></tr> | ||
| + | <tr><td>5-Final Extend</td><td>72<sup>o</sup>C</td><td>15 minutes</td></tr></table> | ||
| + | † Annealing temperature for Phusion is MT +3<sup>o</sup>C <br> | ||
| + | ‡ Extend time is 15-30 seconds per kb; time adjusted accordingly <br> | ||
| + | |||
| + | ===<font color="white">Step 5 - Plasmid DNA Purification=== | ||
| - | |||
We followed Qiagen's QIAPrep Spin Miniprep protocol, as follows: | We followed Qiagen's QIAPrep Spin Miniprep protocol, as follows: | ||
#Pellet cells from 1-1.5mL of liquid culture in a microcentrifuge tube (remove and discard supernatant) | #Pellet cells from 1-1.5mL of liquid culture in a microcentrifuge tube (remove and discard supernatant) | ||
| Line 220: | Line 252: | ||
#Transfer column to clean 1.5mL microcentrifuge tube | #Transfer column to clean 1.5mL microcentrifuge tube | ||
#Add 50µL buffer EB, let stand for 1-5 minutes, and centrifuge for 1 minute (retain flow through) | #Add 50µL buffer EB, let stand for 1-5 minutes, and centrifuge for 1 minute (retain flow through) | ||
| + | <br> | ||
| + | |||
| + | ==<font color="white">Protocols prior to the development of Red/White 3-Antibiotic assembly== | ||
| - | |||
===<font color="white">Competent Cell Transformation=== | ===<font color="white">Competent Cell Transformation=== | ||
| + | |||
<ol> | <ol> | ||
<li>Thaw 20µL of aliquotted cells (DH5α or BL21(DE3)) on ice.</li> | <li>Thaw 20µL of aliquotted cells (DH5α or BL21(DE3)) on ice.</li> | ||
| Line 241: | Line 276: | ||
===<font color="white">Plasmid DNA Purification by Boiling Lysis (Small Scale AKA Miniprep)=== | ===<font color="white">Plasmid DNA Purification by Boiling Lysis (Small Scale AKA Miniprep)=== | ||
| + | |||
<ol> | <ol> | ||
<li>Aseptically transfer 1.5mL of each overnight culture to a 1.5mL microcentrifuge tube (MCT) and pellet the cells by centrifugation in a benchtop microcentrifuge (2min at 13000RPM)</li> | <li>Aseptically transfer 1.5mL of each overnight culture to a 1.5mL microcentrifuge tube (MCT) and pellet the cells by centrifugation in a benchtop microcentrifuge (2min at 13000RPM)</li> | ||
| Line 258: | Line 294: | ||
===<font color="white">Restriction of Plasmid DNA (pDNA)=== | ===<font color="white">Restriction of Plasmid DNA (pDNA)=== | ||
| + | |||
<ol> | <ol> | ||
<li>In a 1.5mL microcentrifuge tube, add MilliQ H<sub>2</sub>O to final volume of 20µL, 2µL of restriction enzyme buffer, 2µL of plasmid DNA, and 0.25µL of each restriction enxyme.</li> | <li>In a 1.5mL microcentrifuge tube, add MilliQ H<sub>2</sub>O to final volume of 20µL, 2µL of restriction enzyme buffer, 2µL of plasmid DNA, and 0.25µL of each restriction enxyme.</li> | ||
| Line 265: | Line 302: | ||
</ol> | </ol> | ||
| - | ==Additional Protocols== | + | <br> |
| + | ==<font color="white">Additional Protocols== | ||
| + | |||
===<font color="white">Overexpression=== | ===<font color="white">Overexpression=== | ||
| + | |||
<ol> | <ol> | ||
<li>Inoculate two 5mL overnight cultures in LB media containing the appropriate antibiotic corresponding to the resistance of the plasmid backbone on which the gene is located; incubated at at 37<sup>o</sup>C in shaker.</li> | <li>Inoculate two 5mL overnight cultures in LB media containing the appropriate antibiotic corresponding to the resistance of the plasmid backbone on which the gene is located; incubated at at 37<sup>o</sup>C in shaker.</li> | ||
<li>Transfer the two 5mL cultures into an 2000mL Erlenmeyer flask containing 500mL of LB media containing the same antibiotic.</li> | <li>Transfer the two 5mL cultures into an 2000mL Erlenmeyer flask containing 500mL of LB media containing the same antibiotic.</li> | ||
| - | <li>Measure and record initial read OD<sub>600</sub> reading (Time = 0) blanking against the | + | <li>Measure and record initial read OD<sub>600</sub> reading (Time = 0) blanking against the uninoculated LB media.</li> |
<li>Begin incubation at 37<sup>o</sup>C in shaker.</li> | <li>Begin incubation at 37<sup>o</sup>C in shaker.</li> | ||
<li>Measure and record another OD<sub>600</sub> reading 1 hour after (Time = 1) and continue record OD<sub>600</sub> every 30 minutes (T + 0.5) until OD<sub>600</sub> = 0.600 is reached.</li> | <li>Measure and record another OD<sub>600</sub> reading 1 hour after (Time = 1) and continue record OD<sub>600</sub> every 30 minutes (T + 0.5) until OD<sub>600</sub> = 0.600 is reached.</li> | ||
| Line 278: | Line 318: | ||
</ol> | </ol> | ||
| - | ===<font color="white"> | + | ===<font color="white">Plasmid DNA Purification by Alkaline Lysis (Large Scale AKA Maxiprep)=== |
| + | |||
<ol> | <ol> | ||
| - | <li>Grow up a 500mL overnight culture in LB media containing the appropriate antibiotic corresponding to the | + | <li>Grow up a 500mL overnight culture in LB media containing the appropriate antibiotic corresponding to the resistance of the plasmid.</li> |
<li>Centrifuge the cells at 5000 rpm for 10 minutes, discard the supernatant, transfer pellet to a 50mL falcon tube, record the weight of the pellet, and store at -20<sup>o</sup>C. ( 1- 2.5g cell pellets are expected)</li> | <li>Centrifuge the cells at 5000 rpm for 10 minutes, discard the supernatant, transfer pellet to a 50mL falcon tube, record the weight of the pellet, and store at -20<sup>o</sup>C. ( 1- 2.5g cell pellets are expected)</li> | ||
<li>Resuspend the cell pellet in 6mL Alkaline Lysis Solution I (ALS1). Vortex carefully and slowly; may use a clean glass stir rod. Add 20µL of 1mg/mL RNase A.</li> | <li>Resuspend the cell pellet in 6mL Alkaline Lysis Solution I (ALS1). Vortex carefully and slowly; may use a clean glass stir rod. Add 20µL of 1mg/mL RNase A.</li> | ||
| Line 308: | Line 349: | ||
<i>RNase A:</i> 1mg/mL in 20mM Tris-HCl, pH 8.0 | <i>RNase A:</i> 1mg/mL in 20mM Tris-HCl, pH 8.0 | ||
<i>Lysozyme:</i> 10mg/mL in 20mM Tris-HCl, pH 8.0 | <i>Lysozyme:</i> 10mg/mL in 20mM Tris-HCl, pH 8.0 | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
 "
"