Project/PlcR-PapR Fusion Protein/
From 2010.igem.org
(→PlcR-PapR Fusion Quorum Peptide - BBa_K354001) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == PlcR-PapR Fusion Quorum Peptide == | + | {{Template:WITS-South_Africa_Main_Menu_19_07}} |
| + | {{Template:WITS-South_Africa_Main_Menu_0910}} | ||
| + | |||
| + | == PlcR-PapR Fusion Quorum Peptide - [http://partsregistry.org/Part:BBa_K354001 BBa_K354001] == | ||
=== Background === | === Background === | ||
| - | + | So as to enact sufficient comms throughout the molecular milieu that comprises the space between, bacteria have developed an ingenious method of binary communication - quorum sensing. Each species of bacteria posses their own unique quorum molecule which acts as a signal to their presence within a population. Since most actions performed by bacteria are done so in huddle formation, the mechanism of quorum sensing allows the local population density to be gauged. Once enough players have joined the ring, a group effort is set into motion and the coordinated whole sets about on their duty. | |
<br /> | <br /> | ||
<br /> | <br /> | ||
| - | + | As synthetic biologists, we have taken a cavalier leap and have ‘hijacked’ a system of quorum sensing from the groups of Gram-positive bacteria ''Bacillus cereus'' and ''Bacillus thruingiensis''. This quorum system is comprised of the transmitter-receiver effectors: the PlcR and PapR quorum peptides and the PlcR promoter which is ‘activated’ by this protein pair [1]. Observable most clearly in ''Bacillus anthracis'', the PlcR regulon is a sequence that controls the transcription of a host of downstream genes via the positive effects exerted by the PlcR and PapR proteins [2]. | |
<br /> | <br /> | ||
<br /> | <br /> | ||
| - | Based on this finding, we have designed a fusion protein that expresses both the PlcR and PapR proteins simultaneously as an amalgamated single. It has been elucidated further that the fusion protein binds to a region within the promoter known as the PlcR box. For more information on the specific interactions between the regulatory proteins and the PlcR promoter, please click on over to the [[PlcR_Promoter]] description and annotation. | + | Quite simply, both PlcR and PapR are synthesized within the bacteria as propeptides, exported out of the cell via the secA pathway, cleaved by an extracellular protease and re-imported into the cell whereby they affect their transcriptional pressure on the PlcR regulon. However, as spoken by a fictitious frenzied scientist; ‘Nature finds a way’. It has been observed in ''Bacillus anthracis'' that the PlcR and PapR proteins are found in the form of a combined fusion protein the performs the same function as the individual proteins combined [2]. |
| + | <br /> | ||
| + | <br /> | ||
| + | Based on this finding, we have designed a fusion protein that expresses both the PlcR and PapR proteins simultaneously as an amalgamated single. It has been elucidated further that the fusion protein binds to a region within the promoter known as the PlcR box. For more information on the specific interactions between the regulatory proteins and the PlcR promoter, please click on over to the [[PlcR_Promoter]] description and annotation. So as to ensure that the ectopic protein performs well in its adopted environment, we have codon optimised the PlcR-PapR fusion sequence to maximise translational efficacy in ''Lactobacillus gasseri'' [3]. | ||
<br /> | <br /> | ||
=== Sequence Developement and Annotation === | === Sequence Developement and Annotation === | ||
<br /> | <br /> | ||
| + | The PlcR-PapR fusion peptide gene was delivered within the pMK plasmid (below). <br /> | ||
| + | [[Image:PlcR-PapR_in_recieved_plasmid.JPG]] <br /> <br /> | ||
| + | |||
| + | Once digested with EcoR1 and Pst1 restriction endonucleases, the PlcR-PapR fragment was gel extracted and purified in preparation for ligation into PSB1C3. The digest yielded debris fragments, of which the fragment corresponding to the PlcR-PapR fusion peptide was clearly visible in Lane 3(1063bp). <br /> | ||
| + | [[Image:PlcR-PapR_digest.JPG]] <br /> <br /> | ||
| + | |||
| + | In order to prepare the PSB1C3 backbone for the insertion of PlcR-PapR, it was linearised via the digestion of EcoR1 and Pst1. Once gel extracted and purified, the PlcR-PapR insert was ligated into the plasmid and redigested with EcoR1 and Pst1 in order to confirm the successful ligation (below). Lanes 6-8 display three positive clones of the insert with the distinct 1063bp band visible. Lane 5 houses the control - digested pMk-PlcR-PapR. The correlation between control lanes and positive clones identifies the presence of the same 1063bp band. <br /> | ||
| + | [[Image:PlcR-PapR_in_PSB1C3.JPG]] <br /> <br /> | ||
| + | |||
| + | The plasmid map of the PlcR-PapR insert within PSB1C3 is shown along with all appropriate annotations and illustrations [4]. <br /> | ||
| + | [[Image:PlcR-PapR_in_PSB1C3._plasmid_map.JPG.JPG ]] <br /> <br /> | ||
| + | |||
=== Testing and Validation === | === Testing and Validation === | ||
<br /> | <br /> | ||
=== References === | === References === | ||
| + | [1] Slamti L. & Lereclus D. (2002), A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the ''Bacillus cereus'' group. The European Molecular Biology Organisation Journal 21: 4550-4559. | ||
| + | <br /> | ||
| + | [2] Pomerantsev A.P., Pomerantseva O.M. & Leppla S.H. (2004), A Spontaneous Translational Fusion of ''Bacillus cereus'' PlcR and PapR Activates Transcription of PlcR-Dependant Genes in ''Bacillus anthracis'' via Binding with a Specific Palindromic Sequence. Infection and Immunity 72:5814-5823. | ||
| + | <br /> | ||
| + | [3] Optimizer, http://genomes.urv.es/OPTIMIZER/, Last accessed September 29, 2010 | ||
| + | [4] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za. | ||
<br /> | <br /> | ||
Latest revision as of 20:09, 25 October 2010

Contents |
PlcR-PapR Fusion Quorum Peptide - [http://partsregistry.org/Part:BBa_K354001 BBa_K354001]
Background
So as to enact sufficient comms throughout the molecular milieu that comprises the space between, bacteria have developed an ingenious method of binary communication - quorum sensing. Each species of bacteria posses their own unique quorum molecule which acts as a signal to their presence within a population. Since most actions performed by bacteria are done so in huddle formation, the mechanism of quorum sensing allows the local population density to be gauged. Once enough players have joined the ring, a group effort is set into motion and the coordinated whole sets about on their duty.
As synthetic biologists, we have taken a cavalier leap and have ‘hijacked’ a system of quorum sensing from the groups of Gram-positive bacteria Bacillus cereus and Bacillus thruingiensis. This quorum system is comprised of the transmitter-receiver effectors: the PlcR and PapR quorum peptides and the PlcR promoter which is ‘activated’ by this protein pair [1]. Observable most clearly in Bacillus anthracis, the PlcR regulon is a sequence that controls the transcription of a host of downstream genes via the positive effects exerted by the PlcR and PapR proteins [2].
Quite simply, both PlcR and PapR are synthesized within the bacteria as propeptides, exported out of the cell via the secA pathway, cleaved by an extracellular protease and re-imported into the cell whereby they affect their transcriptional pressure on the PlcR regulon. However, as spoken by a fictitious frenzied scientist; ‘Nature finds a way’. It has been observed in Bacillus anthracis that the PlcR and PapR proteins are found in the form of a combined fusion protein the performs the same function as the individual proteins combined [2].
Based on this finding, we have designed a fusion protein that expresses both the PlcR and PapR proteins simultaneously as an amalgamated single. It has been elucidated further that the fusion protein binds to a region within the promoter known as the PlcR box. For more information on the specific interactions between the regulatory proteins and the PlcR promoter, please click on over to the PlcR_Promoter description and annotation. So as to ensure that the ectopic protein performs well in its adopted environment, we have codon optimised the PlcR-PapR fusion sequence to maximise translational efficacy in Lactobacillus gasseri [3].
Sequence Developement and Annotation
The PlcR-PapR fusion peptide gene was delivered within the pMK plasmid (below).
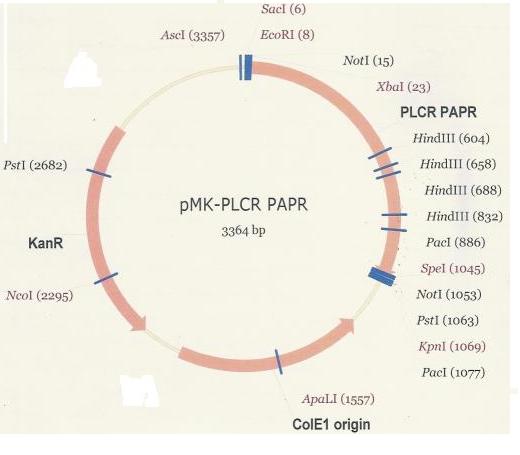
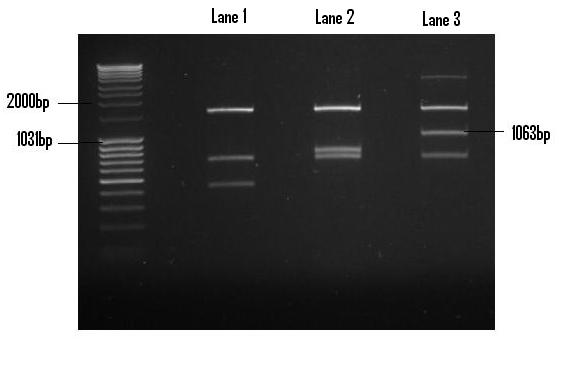
Once digested with EcoR1 and Pst1 restriction endonucleases, the PlcR-PapR fragment was gel extracted and purified in preparation for ligation into PSB1C3. The digest yielded debris fragments, of which the fragment corresponding to the PlcR-PapR fusion peptide was clearly visible in Lane 3(1063bp).
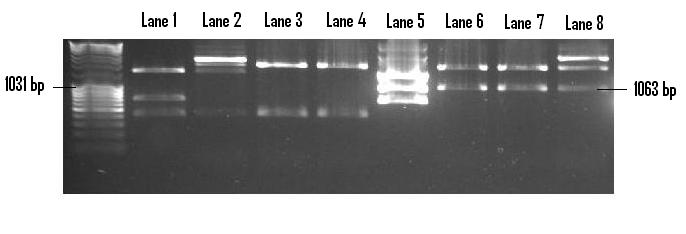
In order to prepare the PSB1C3 backbone for the insertion of PlcR-PapR, it was linearised via the digestion of EcoR1 and Pst1. Once gel extracted and purified, the PlcR-PapR insert was ligated into the plasmid and redigested with EcoR1 and Pst1 in order to confirm the successful ligation (below). Lanes 6-8 display three positive clones of the insert with the distinct 1063bp band visible. Lane 5 houses the control - digested pMk-PlcR-PapR. The correlation between control lanes and positive clones identifies the presence of the same 1063bp band.
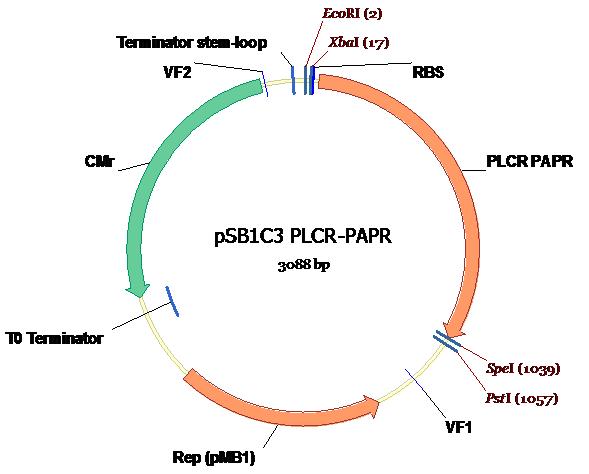
The plasmid map of the PlcR-PapR insert within PSB1C3 is shown along with all appropriate annotations and illustrations [4].
Testing and Validation
References
[1] Slamti L. & Lereclus D. (2002), A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. The European Molecular Biology Organisation Journal 21: 4550-4559.
[2] Pomerantsev A.P., Pomerantseva O.M. & Leppla S.H. (2004), A Spontaneous Translational Fusion of Bacillus cereus PlcR and PapR Activates Transcription of PlcR-Dependant Genes in Bacillus anthracis via Binding with a Specific Palindromic Sequence. Infection and Immunity 72:5814-5823.
[3] Optimizer, http://genomes.urv.es/OPTIMIZER/, Last accessed September 29, 2010
[4] Marco Weinberg, PhD. Antiviral Gene Therapy Research Unit, Department of Molecular Medicine and Haematology, University of the Witwatersrand Medical School, 7 York Rd. Parktown 2193 SOUTH AFRICA, marc.weinberg@wits.ac.za.
 "
"